| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,439,921 – 3,440,011 |
| Length | 90 |
| Max. P | 0.528337 |

| Location | 3,439,921 – 3,440,011 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 52.26 |
| Shannon entropy | 0.85726 |
| G+C content | 0.34714 |
| Mean single sequence MFE | -18.94 |
| Consensus MFE | -3.80 |
| Energy contribution | -5.38 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.528337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3439921 90 - 21146708 --------CCCCUUUUAAUAUCUUAAAUAGAGGAUUGAGCAUAAUCAAUCUUUUAU------AGGGCAAUGUAGCCCAAACAAACACAUU----CUGAAUUGAAAGGA --------..(((((((((.((....((((((((((((......))))))))))))------.((((......)))).............----..))))))))))). ( -29.10, z-score = -5.37, R) >droAna3.scaffold_13266 9961132 77 - 19884421 ---------------------------AAUUUCCUUGAAUCUUACAUAUAUUCUUGU-AAUCAAUCUU-UACGUGUUAGUUUCCCUUCUA--AGGCCGAUUGAAAGGA ---------------------------....(((((.....(((((........)))-))((((((..-...((.((((........)))--).)).))))))))))) ( -15.10, z-score = -2.16, R) >droYak2.chr2L 16118237 96 - 22324452 --------CCCCCUUUAAUAUCAAAAAUAGAGGAUUGGGCAUAAUCAAUCUUUUAUACUCGUAAGUCUAUGUAGCCCAGGCACACACAUA----CUAAAUUGAAAGGA --------..((.((((((.......((((((((((((......))))))))))))....(((.((...(((.((....))))).)).))----)...)))))).)). ( -21.00, z-score = -1.81, R) >droEre2.scaffold_4929 7483577 89 + 26641161 --------CCAUCUUCGAUAUUAAAAAUAGAGGAUUGGGCAUAAUCAAUCUUUCAA------AGGUCUAUGUAG-CCAGGCACACACAUA----CUGAAUUGAAAGGA --------((((((((.............))))).)))..........((((((((------.(((.(.(((.(-(...))..))).).)----))...)))))))). ( -15.22, z-score = -0.15, R) >droSim1.chr2R 2165439 102 - 19596830 CCCCCAUUCCCCAUUCCAUAUCUUAAAUAGAGGAUUGGGCAUAAUCAAUCUUUUAU------GGGGCAAUGUAGCCCAGACACACACACACACUCUGAAUUGAAAGGA ...((.(((...((((..........((((((((((((......))))))))))))------.((((......))))...................)))).))).)). ( -22.90, z-score = -0.61, R) >droVir3.scaffold_12875 6120406 93 + 20611582 ------------AAUUCACUUCAUACGAAGUUCAUUAAGUUUGGAUAUUCCUUUUAA-AGGCAAGCUUAUACAUAUGACAAAAAUAUAUA--UUUUAAAGGUACAGUA ------------.....(((((....)))))((((((((((((......(((.....-))))))))))).....))))............--................ ( -10.30, z-score = 0.10, R) >consensus ________CCCCAUUUAAUAUCAUAAAUAGAGGAUUGAGCAUAAUCAAUCUUUUAU______AAGCCUAUGUAGCCCAGACACACACAUA____CUGAAUUGAAAGGA ...........................(((((((((((......)))))))))))..................................................... ( -3.80 = -5.38 + 1.58)

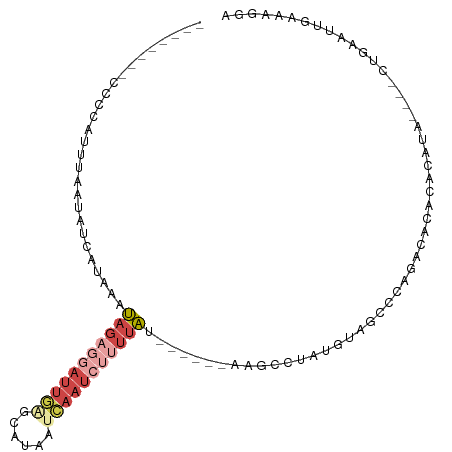

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:49 2011