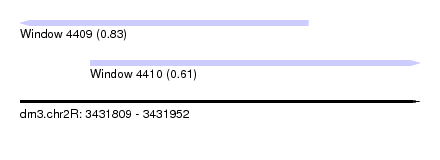
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,431,809 – 3,431,952 |
| Length | 143 |
| Max. P | 0.828880 |

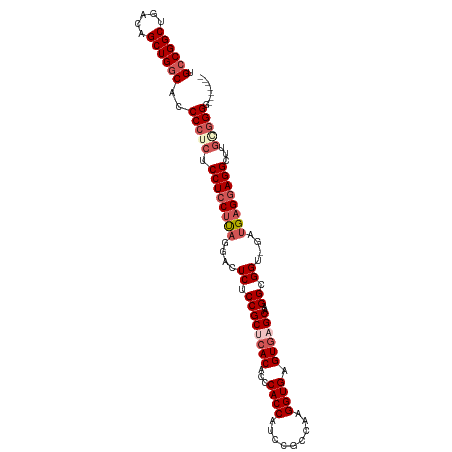
| Location | 3,431,809 – 3,431,912 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.13 |
| Shannon entropy | 0.28384 |
| G+C content | 0.63643 |
| Mean single sequence MFE | -45.38 |
| Consensus MFE | -37.66 |
| Energy contribution | -39.54 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.828880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3431809 103 - 21146708 UGCCGGCUGACAGCUGGCACCCCUCUCCUCCUUUGGACUCUCCGCUCACACCCACCAUCCGCCAAGGUGAGUGAGCAAGGCGGU-GAUGAGAAGGGUUGGCGGG------- (((((((.....))))))).(((.((..((((((....((.((((((.(((.((((.........)))).))).)...))))).-))...))))))..)).)))------- ( -41.20, z-score = -0.40, R) >droEre2.scaffold_4929 7474038 105 + 26641161 UGCCGGCUGACAGCUGGCUACCCUCCCCUCCACA-GACUCUCCGCCCAC---CACCCCC-GUCAAGGUGAGUGAGCAAGGCGGU-GAUGAGGAGGCUUGAGGGGGUACCAU .((((((.....))))))..(((((.(((((.((-...((.((((((((---((((...-.....)))).))).....))))).-)))).)))))...)))))........ ( -47.60, z-score = -1.80, R) >droYak2.chr2L 16108642 106 - 22324452 UGCCGGCUGACAGCUGGCUGCCCUCCCCUCCUCA-GAAUCUCCGCCCAC---CACCCCCUGCCAAGGUGAGUUAGCAAGGCGGU-GAUGAGGAGGCUUGCGGGGGUACCAU .((((((.....))))))((((((((((((((((-...((.((((((((---(............)))).(....)..))))).-))))))))))...).))))))).... ( -48.30, z-score = -1.68, R) >droSec1.super_1 1094136 102 - 14215200 UGCCGGCUGACAGCUGGCAUCCCUCUCCUCCUUAGGACUCUCCGCUCACACCCACCAUCCGCCAAGGUGAGUGAGCAAGGCGGU-UAUGAGGAGGGUUGCGGG-------- (((((((.....))))))).(((.((((((((((.((((((..((((((...((((.........)))).))))))..)).)))-).)))))))))..).)))-------- ( -46.30, z-score = -2.24, R) >droSim1.chr2R 2156348 104 - 19596830 UGCCGGCUGACAGCUGACACCCCUCUCCUCCUUAGGACUCUCCGCUCACACCCACCAUCCGCCAAGGUGAGUGAGCAAGGCGGUAGAUGAGGAGGUUUUGGGGG------- ((.((((.....)))).))((((...((((((((..(((((..((((((...((((.........)))).))))))..)).)))...))))))))....)))).------- ( -43.50, z-score = -1.45, R) >consensus UGCCGGCUGACAGCUGGCACCCCUCUCCUCCUUAGGACUCUCCGCUCACACCCACCAUCCGCCAAGGUGAGUGAGCAAGGCGGU_GAUGAGGAGGCUUGCGGGG_______ .((((((.....))))))..(((((.((((((((....((.((((((((...((((.........)))).))))))..)).))....))))))))...)))))........ (-37.66 = -39.54 + 1.88)

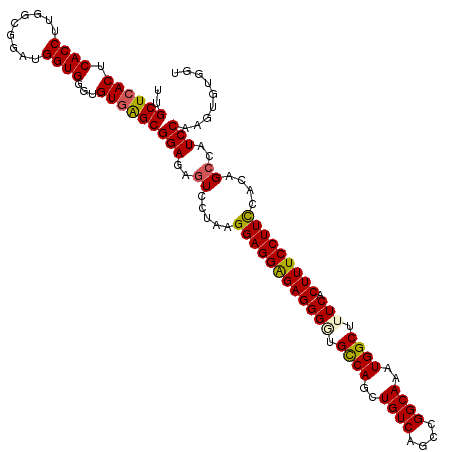
| Location | 3,431,834 – 3,431,952 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 89.97 |
| Shannon entropy | 0.17538 |
| G+C content | 0.58021 |
| Mean single sequence MFE | -48.50 |
| Consensus MFE | -41.50 |
| Energy contribution | -41.42 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.608533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3431834 118 + 21146708 UUGCUCACUCACCUUGGCGGAUGGUGGGUGUGAGCGGAGAGUCCAAAGGAGGAGAGGGGUGCCAGCUGUCAGCCGGCAAAUGGCUUUCACUUUCCUUCCACAGCCAUCCAAGCGUGGU ........(((((((((......((((.((((...((.....))...((((((((((((.((((..((((....))))..)))).))).))))))))))))).))))))))).)))). ( -47.20, z-score = -0.82, R) >droEre2.scaffold_4929 7474070 113 - 26641161 UUGCUCACUCACCUUGACGGG-GGUG---GUGGGCGGAGAGUC-UGUGGAGGGGAGGGUAGCCAGCUGUCAGCCGGCAAAUGGCUUUCACUUUCCUUCCACAGCCAUCCAGGUGUGGU ..((((((.(((((......)-))))---))))))(((..(.(-(((((((((((((..(((((..((((....))))..)))))....)))))))))))))))..)))......... ( -59.30, z-score = -4.05, R) >droYak2.chr2L 16108674 114 + 22324452 UUGCUAACUCACCUUGGCAGGGGGUG---GUGGGCGGAGAUUC-UGAGGAGGGGAGGGCAGCCAGCUGUCAGCCGGCAAAUGGCUUUCACUUUCCUUUCACAGCAAUCCGAGUGUGGU ..(((.((.((((((.....))))))---)).)))(((....(-((.((((((((((..(((((..((((....))))..)))))....)))))))))).)))...)))......... ( -44.50, z-score = -0.76, R) >droSec1.super_1 1094160 118 + 14215200 UUGCUCACUCACCUUGGCGGAUGGUGGGUGUGAGCGGAGAGUCCUAAGGAGGAGAGGGAUGCCAGCUGUCAGCCGGCAAAUGGCUUUCACUUUCCUUCCACAGCCAUCCAAGUGUGGU ........(((((((((......((((.((((...((.....))...((((((((((((.((((..((((....))))..)))).))).))))))))))))).))))))))).)))). ( -47.50, z-score = -1.18, R) >droSim1.chr2R 2156374 118 + 19596830 UUGCUCACUCACCUUGGCGGAUGGUGGGUGUGAGCGGAGAGUCCUAAGGAGGAGAGGGGUGUCAGCUGUCAGCCGGCAAAUGGCUUUCACUUUCCUUCCACAGCCAUCCAAGUGUGGU ........(((((((((......((((.((((...((.....))...((((((((((((.((((..((((....))))..)))).))).))))))))))))).))))))))).)))). ( -44.00, z-score = -0.29, R) >consensus UUGCUCACUCACCUUGGCGGAUGGUGGGUGUGAGCGGAGAGUCCUAAGGAGGAGAGGGGUGCCAGCUGUCAGCCGGCAAAUGGCUUUCACUUUCCUUCCACAGCCAUCCAAGUGUGGU ..((((((.((((.........))))...))))))(((..((.....((((((((((((.((((..((((....))))..)))).))).)))))))))....))..)))......... (-41.50 = -41.42 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:48 2011