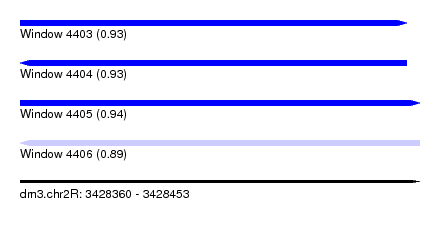
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,428,360 – 3,428,453 |
| Length | 93 |
| Max. P | 0.944418 |

| Location | 3,428,360 – 3,428,450 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 61.70 |
| Shannon entropy | 0.67945 |
| G+C content | 0.34432 |
| Mean single sequence MFE | -16.46 |
| Consensus MFE | -7.88 |
| Energy contribution | -7.60 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.930941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
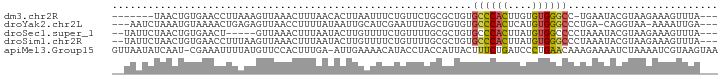
>dm3.chr2R 3428360 90 + 21146708 -------UAACUGUGAACCUUAAAGUUAAACUUUAACACUUAAUUUCUGUUCUGCGCUGUGCCCACUUGUGUGGGCC-UGAAUACGUAAGAAAGUUUA--- -------.(((((((....((((((.....)))))))))..........(((((((.(((((((((....)))))).-...)))))).))))))))..--- ( -20.10, z-score = -1.83, R) >droYak2.chr2L 16105444 93 + 22324452 ---AAUCUAAAUGUAAAACUGAGAGUUAACCUUUUAUAAUUGCAUCGAAUUUAGCUGUGUGCCCACUCAUGUGGGCCCUGA-CAGGUAA-AAAAUUGA--- ---.......((((((...((((((.....))))))...))))))....((((.((((..((((((....))))))....)-))).)))-).......--- ( -23.90, z-score = -2.91, R) >droSec1.super_1 1090676 91 + 14215200 --UAUUCUAACUGUGAACU-----GUUAAACUUUAAUACUUGUUUUCUGUUUUGCGCUGUGCCCACUUAUGUGGCCCCUAAAUACGUAAGAAAGUUUA--- --......(((.(....).-----)))(((((((..(((.(((((........((.....))((((....)))).....))))).)))..))))))).--- ( -13.70, z-score = -0.76, R) >droSim1.chr2R 2152675 96 + 19596830 --UAUUCUAACUGUGAACCUUUAAGUUAAACUUUAAUACUUGUUUUCUGUUUUGCGCUGUGCCCACUUAUGUGGGCCCUAAAUACGUAAGAAAGUUUA--- --........................((((((((..(((.(((((.............(.((((((....)))))).).))))).)))..))))))))--- ( -18.84, z-score = -1.64, R) >apiMel3.Group15 3648877 99 - 7856270 GUUAAUAUCAAU-CGAAAUUUUAUGUUCCACUUUGA-AUUGAAAACAUACCUACCAUUACUUUCUGAUCCCUGAACAAAGAAAAUCUAAAAUCGUAAGUAA .......(((((-((((..............)))).-)))))..............((((((..((((....((..........))....)))).)))))) ( -5.74, z-score = 1.18, R) >consensus __UAAUCUAACUGUGAACCUUAAAGUUAAACUUUAAUACUUGAUUUCUGUUUUGCGCUGUGCCCACUUAUGUGGGCCCUGAAUACGUAAGAAAGUUUA___ ............................................................((((((....))))))......................... ( -7.88 = -7.60 + -0.28)
| Location | 3,428,360 – 3,428,450 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 61.70 |
| Shannon entropy | 0.67945 |
| G+C content | 0.34432 |
| Mean single sequence MFE | -17.28 |
| Consensus MFE | -8.33 |
| Energy contribution | -8.05 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.930941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3428360 90 - 21146708 ---UAAACUUUCUUACGUAUUCA-GGCCCACACAAGUGGGCACAGCGCAGAACAGAAAUUAAGUGUUAAAGUUUAACUUUAAGGUUCACAGUUA------- ---..((((......(((.....-.((((((....))))))...)))..((((.....(((((.((((.....))))))))).))))..)))).------- ( -19.50, z-score = -1.85, R) >droYak2.chr2L 16105444 93 - 22324452 ---UCAAUUUU-UUACCUG-UCAGGGCCCACAUGAGUGGGCACACAGCUAAAUUCGAUGCAAUUAUAAAAGGUUAACUCUCAGUUUUACAUUUAGAUU--- ---.......(-(((.(((-(....((((((....))))))..)))).)))).............((((.(...(((.....)))...).))))....--- ( -17.20, z-score = -0.79, R) >droSec1.super_1 1090676 91 - 14215200 ---UAAACUUUCUUACGUAUUUAGGGGCCACAUAAGUGGGCACAGCGCAAAACAGAAAACAAGUAUUAAAGUUUAAC-----AGUUCACAGUUAGAAUA-- ---((((((((..(((...........((((....))))((.....))..............)))..))))))))..-----.((((.......)))).-- ( -15.70, z-score = -1.52, R) >droSim1.chr2R 2152675 96 - 19596830 ---UAAACUUUCUUACGUAUUUAGGGCCCACAUAAGUGGGCACAGCGCAAAACAGAAAACAAGUAUUAAAGUUUAACUUAAAGGUUCACAGUUAGAAUA-- ---((((((((..(((.........((((((....))))))....(........).......)))..))))))))........((((.......)))).-- ( -19.60, z-score = -2.09, R) >apiMel3.Group15 3648877 99 + 7856270 UUACUUACGAUUUUAGAUUUUCUUUGUUCAGGGAUCAGAAAGUAAUGGUAGGUAUGUUUUCAAU-UCAAAGUGGAACAUAAAAUUUCG-AUUGAUAUUAAC .(((((((.(((....(((((((..(((....))).)))))))))).))))))).....(((((-(.(((.((.....))...))).)-)))))....... ( -14.40, z-score = 0.29, R) >consensus ___UAAACUUUCUUACGUAUUCAGGGCCCACAUAAGUGGGCACAGCGCAAAACAGAAAACAAGUAUUAAAGUUUAACUUAAAGGUUCACAGUUAGAAUA__ ........(((((............((((((....))))))............)))))........................................... ( -8.33 = -8.05 + -0.28)
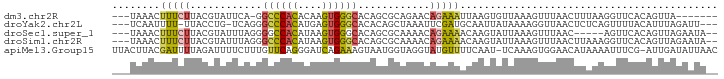
| Location | 3,428,360 – 3,428,453 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 51.65 |
| Shannon entropy | 0.92171 |
| G+C content | 0.33842 |
| Mean single sequence MFE | -15.83 |
| Consensus MFE | -5.84 |
| Energy contribution | -5.90 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
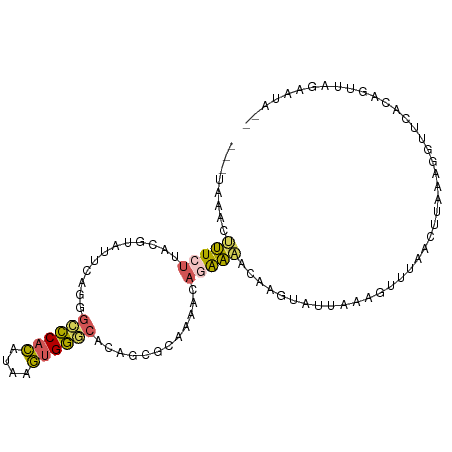
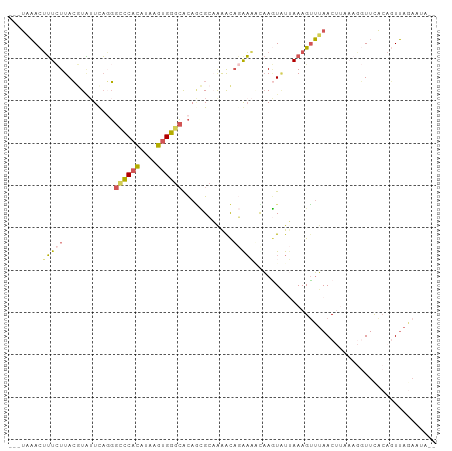
>dm3.chr2R 3428360 93 + 21146708 -UAACUGU-GAACCUUAAAGUUAAACUUUAACACUUAAUUUCUGUUCUGCGCUGUGCCCACUUGUGUGGGCC-UGAAUACGUAAGAAAGUUUAUCA------ -.....((-((((.((((((.....)))))).......(((((....((((.(((((((((....)))))).-...))))))))))))))))))..------ ( -21.10, z-score = -1.98, R) >droYak2.chr2L 16105448 92 + 22324452 -UAAAUGU-AAAACUGAGAGUUAACCUUUUAUAAUUGCAUCGAAUUUAGCUGUGUGCCCACUCAUGUGGGCCCUGA-CAGGUAA-AAAAUUGAUUA------ -....(((-((...((((((.....))))))...)))))((((.((((.((((..((((((....))))))....)-))).)))-)...))))...------ ( -24.10, z-score = -3.01, R) >droSec1.super_1 1090680 90 + 14215200 CUAACUGU-GAACU-----GUUAAACUUUAAUACUUGUUUUCUGUUUUGCGCUGUGCCCACUUAUGUGGCCCCUAAAUACGUAAGAAAGUUUAUCA------ ........-.....-----(.((((((((..(((.(((((........((.....))((((....)))).....))))).)))..)))))))).).------ ( -14.40, z-score = -0.79, R) >droSim1.chr2R 2152679 95 + 19596830 CUAACUGU-GAACCUUUAAGUUAAACUUUAAUACUUGUUUUCUGUUUUGCGCUGUGCCCACUUAUGUGGGCCCUAAAUACGUAAGAAAGUUUAUCA------ ........-..........(.((((((((..(((.(((((.............(.((((((....)))))).).))))).)))..)))))))).).------ ( -20.04, z-score = -1.96, R) >droGri2.scaffold_15245 10565288 74 + 18325388 ---------------ACAAAGAUGACUUUGAAAUUUAUUGCAUA------AACAAAUGCCUGUAAAGUGUCGGAAAAUUUAUAAGAAAUCAAGCA------- ---------------....((((...(((((..(((((.((((.------.....))))..)))))...)))))..))))...............------- ( -9.10, z-score = -0.01, R) >apiMel3.Group15 3648881 101 - 7856270 AUAUCAAUCGAAAUUUUAUGUUCCACUUUGA-AUUGAAAACAUACCUACCAUUACUUUCUGAUCCCUGAACAAAGAAAAUCUAAAAUCGUAAGUAACCAUAA ...(((((((((..............)))).-)))))..............((((((..((((....((..........))....)))).))))))...... ( -6.24, z-score = 0.72, R) >consensus _UAACUGU_GAACCUUAAAGUUAAACUUUAAUACUUAUUUCCUAUUUUGCGCUGUGCCCACUUAUGUGGGCCCUAAAUACGUAAGAAAGUUUAUCA______ .......................................................((((((....))))))............................... ( -5.84 = -5.90 + 0.06)
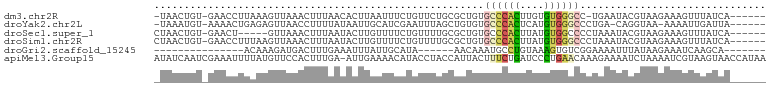
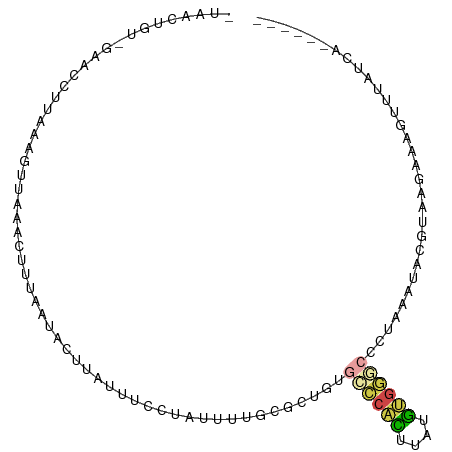
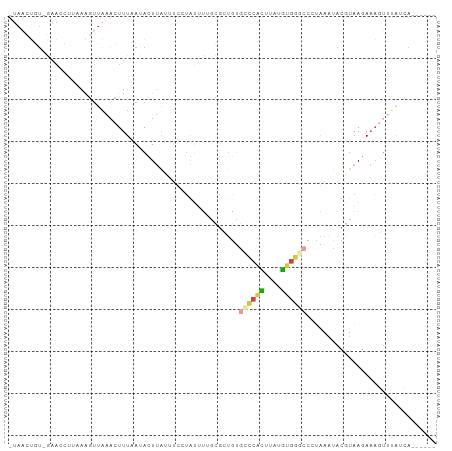
| Location | 3,428,360 – 3,428,453 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 51.65 |
| Shannon entropy | 0.92171 |
| G+C content | 0.33842 |
| Mean single sequence MFE | -15.89 |
| Consensus MFE | -6.19 |
| Energy contribution | -5.97 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3428360 93 - 21146708 ------UGAUAAACUUUCUUACGUAUUCA-GGCCCACACAAGUGGGCACAGCGCAGAACAGAAAUUAAGUGUUAAAGUUUAACUUUAAGGUUC-ACAGUUA- ------.....((((......(((.....-.((((((....))))))...)))..((((.....(((((.((((.....))))))))).))))-..)))).- ( -19.50, z-score = -1.36, R) >droYak2.chr2L 16105448 92 - 22324452 ------UAAUCAAUUUU-UUACCUG-UCAGGGCCCACAUGAGUGGGCACACAGCUAAAUUCGAUGCAAUUAUAAAAGGUUAACUCUCAGUUUU-ACAUUUA- ------.....((((((-(((((..-...))((((((....)))))).....((..........)).....))))))))).............-.......- ( -17.10, z-score = -0.95, R) >droSec1.super_1 1090680 90 - 14215200 ------UGAUAAACUUUCUUACGUAUUUAGGGGCCACAUAAGUGGGCACAGCGCAAAACAGAAAACAAGUAUUAAAGUUUAAC-----AGUUC-ACAGUUAG ------((.((((((((..(((...........((((....))))((.....))..............)))..)))))))).)-----)....-........ ( -15.90, z-score = -1.39, R) >droSim1.chr2R 2152679 95 - 19596830 ------UGAUAAACUUUCUUACGUAUUUAGGGCCCACAUAAGUGGGCACAGCGCAAAACAGAAAACAAGUAUUAAAGUUUAACUUAAAGGUUC-ACAGUUAG ------...((((((((..(((.........((((((....))))))....(........).......)))..))))))))............-........ ( -19.00, z-score = -1.74, R) >droGri2.scaffold_15245 10565288 74 - 18325388 -------UGCUUGAUUUCUUAUAAAUUUUCCGACACUUUACAGGCAUUUGUU------UAUGCAAUAAAUUUCAAAGUCAUCUUUGU--------------- -------((((((...........................)))))).((((.------...)))).......(((((....))))).--------------- ( -7.53, z-score = 0.25, R) >apiMel3.Group15 3648881 101 + 7856270 UUAUGGUUACUUACGAUUUUAGAUUUUCUUUGUUCAGGGAUCAGAAAGUAAUGGUAGGUAUGUUUUCAAU-UCAAAGUGGAACAUAAAAUUUCGAUUGAUAU ((((.(((((((.((((((..((..........))..))))).).))))))).)))).(((((((.((..-......)))))))))................ ( -16.30, z-score = -0.07, R) >consensus ______UGAUAAACUUUCUUACGUAUUCAGGGCCCACAUAAGUGGGCACAGCGCAAAACAGAAAACAAGUAUUAAAGUUUAACUUAAAGGUUC_ACAGUUA_ ...............................((((((....))))))....................................................... ( -6.19 = -5.97 + -0.22)

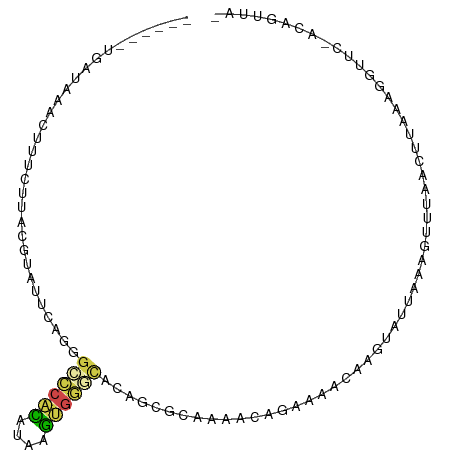
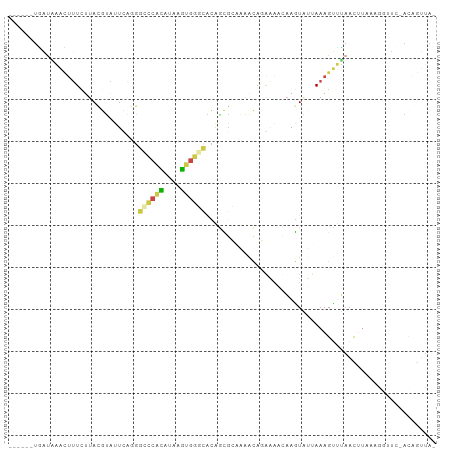
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:45 2011