| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,410,453 – 3,410,561 |
| Length | 108 |
| Max. P | 0.782989 |

| Location | 3,410,453 – 3,410,561 |
|---|---|
| Length | 108 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.69 |
| Shannon entropy | 0.63171 |
| G+C content | 0.51389 |
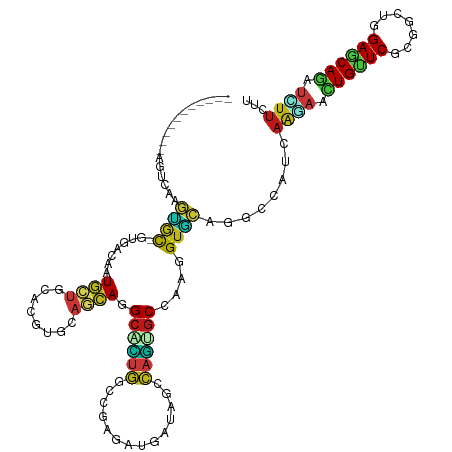
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -16.13 |
| Energy contribution | -15.85 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
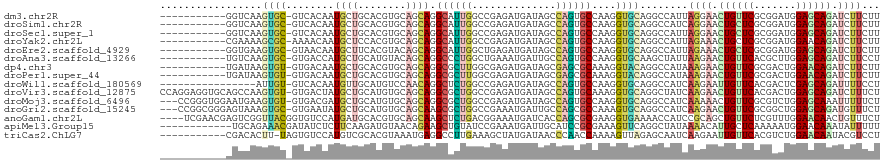
>dm3.chr2R 3410453 108 - 21146708 -----------GGUCAAGUGC-GUCACAAUGCUGCACGUGCAGCAGGCAUUGGCCGAGAUGAUAGCCAGUGCCAAGGUGCAGGCCAUUAGGAACUGUUCGCGGAUGGAGCAGAUCUUCUU -----------((((...(((-..(....((((((....))))))(((((((((..........)))))))))..)..)))))))...((((.((((((.......)))))).))))... ( -45.60, z-score = -2.66, R) >droSim1.chr2R 2131767 108 - 19596830 -----------GGUCAAGUGC-GUCACAAUGCUGCACGUGCAGCAGGCAUUGGCCGAGAUGAUAGCCAGUGCCAAGGUGCAGGCCAUCAGGAACUGCUCGCGGAUGGAGCAGAUCUUCUU -----------((((...(((-..(....((((((....))))))(((((((((..........)))))))))..)..)))))))...((((.((((((.......)))))).))))... ( -48.10, z-score = -2.69, R) >droSec1.super_1 1067354 108 - 14215200 -----------GGUCAAGUGC-GUCACAAUGCUGCACGUGCAGCAGGCAUUGGCCGAGAUGAUAGCCAGUGCCAAGGUGCAGGCCAUUAGGAACUGCUCGCGGAUGGAGCAGAUCUUCUU -----------((((...(((-..(....((((((....))))))(((((((((..........)))))))))..)..)))))))...((((.((((((.......)))))).))))... ( -48.00, z-score = -2.94, R) >droYak2.chr2L 16087337 108 - 22324452 -----------CGAAAAGCGC-AAAACAAUGCUCCACGUGCAGCAGGCAUUGGCCGAGAUGAUAGCCAGUGCCAAGGUGCAGGCCAUUAGAAACUGCUCGCGGAUGGAACAGAUCUUCUU -----------.(((....((-(......)))((((((((.(((((((((((((..........)))))))))..(((....))).........))))))))..)))).......))).. ( -34.10, z-score = -0.68, R) >droEre2.scaffold_4929 7452443 108 + 26641161 -----------GGUGAAGUGC-GUAACAAUGCUUCACGUACAGCAGGCAUUGGCUGAGAUGAUAGCCAGUGCCAAGGUGCAGGCCAUUAGAAACUGCUCGCGGAUGGAGCAGAUCUUCUU -----------.(((((((..-........)))))))((((....(((((((((((......)))))))))))...))))........((((.((((((.......))))))...)))). ( -40.90, z-score = -2.19, R) >droAna3.scaffold_13266 3514514 108 + 19884421 -----------UGUCAAGUGC-GUGACCAUGCUGCAUGUACAGCAGGCCCUGGCUGAAAUGAUUGCCAGUGCCAAGGUGCAAGCUAUUAAGAACUGUUCACGCUUGGAGCAGAUCUUCCU -----------........((-.(((((.(((((......)))))(((.(((((..........))))).)))..))).)).))....((((.((((((.......)))))).))))... ( -36.80, z-score = -0.99, R) >dp4.chr3 7445546 108 + 19779522 -----------UGAUAAGUGU-GUGACAAUGCUGCACGUGCAGCAGGCGCUUGGCGAGAUGAUAGCGAGCGCAAAGGUACAGGCCAUAAAGAACUGUUCGCGACUGGAACAGAUCUUCUU -----------......((.(-(((.(..((((((....)))))).(((((((.(.........))))))))...).)))).))....((((.((((((.......)))))).))))... ( -36.00, z-score = -1.23, R) >droPer1.super_44 98537 108 + 614636 -----------UGAUAAGUGU-GUGACAAUGCUGCACGUGCAGCAGGCGCUUGGCGAGAUGAUAGCGAGCGCAAAGGUACAGGCCAUAAAGAACUGUUCGCGACUGGAACAGAUCUUCUU -----------......((.(-(((.(..((((((....)))))).(((((((.(.........))))))))...).)))).))....((((.((((((.......)))))).))))... ( -36.00, z-score = -1.23, R) >droWil1.scaffold_180569 1300672 103 + 1405142 ----------------AUUGU-GUCACAAUGUUGCAUGUCCAACAGGCUCUGGCCGAGAUGAUAGCCAGUGCCAAGGUGCAGGCCAUCAAGAAUUGUUCACGACUCGAGCAGAUUUUCCU ----------------.(((.-.((....(((((......)))))(((.(((((..........))))).)))..))..)))......((((.((((((.......)))))).))))... ( -27.90, z-score = 0.12, R) >droVir3.scaffold_12875 17554868 119 - 20611582 CCAGGAGGUGCAGCCAAGUGU-GUGACUAUGCUGCAUGUGCAGCAGGCGCUGGCCGAGAUGAUAGCCAGUGCAAAGGUGCAGGCUAUCAAGAACUGUUCACGACUGGAGCAGAUCUUUCU ...((.(((((.....(((..-...))).((((((....)))))).)))))..))((((((((((((.(..(....)..).)))))))).((.((((((.......)))))).)))))). ( -45.50, z-score = -0.93, R) >droMoj3.scaffold_6496 11184165 116 - 26866924 ---CCGGGUGGAAUGAAGUGU-GUGACGAUGCUGCAUGUGCAGCAGGCGCUGGCCGAGAUGAUAGCCAGUGCCAAGGUGCAGGCCAUCAAAAACUGUUCGCGUCUGGAGCAAAUUUUUCU ---((((..(((((..(((..-.(((...((((((....))))))(((((((((..........)))))))))..(((....))).)))...))))))).)..))))............. ( -43.80, z-score = -1.44, R) >droGri2.scaffold_15245 17093373 116 - 18325388 ---CCGGCGGGAGUAAAGUGC-GUGAAUAUGCUGCAUGUGCAGCAAGCGCUGGCCGAAAUGAUUGCCAGCGCCAAAGUGCAGGCCAUCAAGAACUGUUCGCGGCUGGAGCAGAUGUUUCU ---((((((...((.....))-.......((((((....))))))..))))))..((((((((.(((.((((....)))).))).))).....((((((.......))))))..))))). ( -41.40, z-score = 0.03, R) >anoGam1.chr2L 42123502 116 + 48795086 ----UCGAACGAGUCGGUUACGGUGUCCAUGAUGCACGUGCAGCAAGCUCUGACGGAAAUGAUCACCAGCGCGAAGGUGAAAACCAUCCGCAGCUGUUCUCGUUUGGAACAACUGUUUCU ----..(((((((.(((((..((((..(((....(.(((.(((......)))))))..)))..)))).(((.((.(((....))).))))))))))..)))))))(((((....))))). ( -40.80, z-score = -1.87, R) >apiMel3.Group15 5841750 108 + 7856270 ------------UGCAGAAACGAUAUCUCUUCAAGAUGUAACAGAAGCUGUAUCCGAAAUGAUUGCAUCCGCGAAAGUUCAGGCUAUAAAACAUUGCUCAAAAAUGGAACAAAUAUUUUU ------------(((((...(.((((((.....))))))....)...)))))...((((((.(((..(((((....))...(((...........))).......))).))).)))))). ( -16.80, z-score = 0.47, R) >triCas2.ChLG7 11516014 108 - 17478683 -----------CGACACUU-UAGUGUCCAUGUCGCACGUAAAUGAGGCCUUGAAAGCUAUGAUAACCCAACCAAAAGUUAGAGCAAUCAAGAAUUGUUCACGUCUGGAACAAUACGUCCU -----------.(((((..-..)))))......(.(((((........(((((..(((....((((..........)))).)))..)))))..((((((.......))))))))))).). ( -23.80, z-score = -1.27, R) >consensus ___________AGUCAAGUGC_GUGACAAUGCUGCACGUGCAGCAGGCACUGGCCGAGAUGAUAGCCAGUGCCAAGGUGCAGGCCAUCAAGAACUGUUCGCGGCUGGAGCAGAUCUUCUU .............................((((........)))).((((((..............))))))...(((....)))...((((.((((((.......)))))).))))... (-16.13 = -15.85 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:41 2011