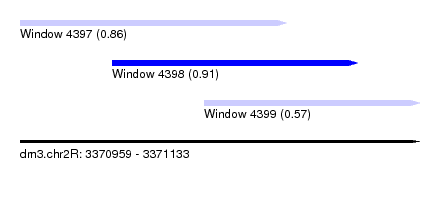
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,370,959 – 3,371,133 |
| Length | 174 |
| Max. P | 0.913592 |

| Location | 3,370,959 – 3,371,075 |
|---|---|
| Length | 116 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.10 |
| Shannon entropy | 0.41558 |
| G+C content | 0.52537 |
| Mean single sequence MFE | -40.13 |
| Consensus MFE | -25.31 |
| Energy contribution | -25.21 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.858038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
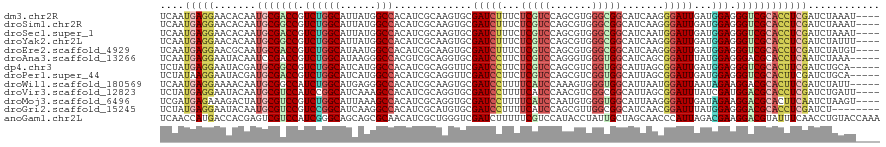
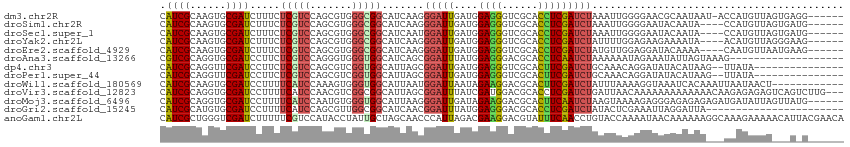
>dm3.chr2R 3370959 116 + 21146708 UCAAUGAGGAACACAAUGCGACCGUCUGGCAUUAUGGCCACAUCGCAAGUGCGAUCUUUCUCGUCCAGCGUGGGCGGCAUCAAGGGAUUGAUGGAGGGUCGCACCUCGAUCUAAAU---- ....(((((.......(((((.....((((......))))..)))))..((((((((((.((((((.....))))))((((((....)))))))))))))))))))))........---- ( -43.30, z-score = -2.18, R) >droSim1.chr2R 2092348 116 + 19596830 UCAAUGAGGAACACAAUGCGGCCGUCUGGCAUUAUGGCCACAUCGCAAGUGCGAUCUUUCUCGUCCAGCGUGGGCGGCAUCAAGGGAUUGAUGGAGGGUCGCACCUCGAUCUAAAU---- ....(((((......(((.((((((........)))))).)))......((((((((((.((((((.....))))))((((((....)))))))))))))))))))))........---- ( -43.40, z-score = -1.95, R) >droSec1.super_1 1028212 116 + 14215200 UCAAUGAGGAACACAAUGCGACCGUCUGGCAUUAUGGCCACAUCGCAAGUGCGAUCUUUCUCGUCCAGCGUGGGCGGCAUCAAUGGAUUGAUGGAGGGUCGCACCUCGAUCUAAAU---- ....(((((.......(((((.....((((......))))..)))))..((((((((((.((((((.....))))))((((((....)))))))))))))))))))))........---- ( -42.50, z-score = -2.02, R) >droYak2.chr2L 16049539 116 + 22324452 UCAAUGAGGAACACAAUGCGGCCGUCUGGCAUUAUGGCCACAUCGCAAGUGCGAUCUUUCUCGUCCAGCGUGGGCGGCAUCAAGGGAUUGAUGGAGGGUCGCACCUCGAUCUAUUU---- ....(((((......(((.((((((........)))))).)))......((((((((((.((((((.....))))))((((((....)))))))))))))))))))))........---- ( -43.40, z-score = -1.85, R) >droEre2.scaffold_4929 7413598 116 - 26641161 UCAAUGAGGAACGCAAUGCGACCGUCUGGCAUAAUGGCCACAUCGCAAGUGCGAUCUUUCUCGUCCAGCGUGGGCGGCAUCAAGGGAUUGAUGGAGGGUCGCACCUCGAUCUAUGU---- ....(((((.......(((((.....((((......))))..)))))..((((((((((.((((((.....))))))((((((....)))))))))))))))))))))........---- ( -43.30, z-score = -1.39, R) >droAna3.scaffold_13266 3476680 115 - 19884421 UCAAUGAGGAAUACAAUCCGACCGUCUGGCAUAAGGGCCACGUCGCAGGUGCGAUCCUUCUCGUCCAGGGUGGGUGGCAUCAGCGGAUUUAUGGAGGGACGCACCUCAAUCUAAA----- .......(((......)))(((....((((......)))).)))(.(((((((.(((((((.((((..((((.....))))...))))....)))))))))))))))........----- ( -39.30, z-score = -0.87, R) >dp4.chr3 7405670 115 - 19779522 UCUAUGAGGAAUACGAUGCGGCCGUCUGGCAUCAUGGCCACAUCGCAGGUUCGAUCCUUCUCGUCCAGCGUCGGUGGCAUUAGCGGAUUGAUGGAGGGUCGCACUUCGAUCUGCA----- ..............((((.((((((........)))))).))))((((((((((((((((((((((.((.............))))).))).)))))))))......))))))).----- ( -42.92, z-score = -0.82, R) >droPer1.super_44 58559 115 - 614636 UCUAUAAGGAAUACGAUGCGACCGUCUGGCAUCAUGGCCACAUCGCAGGUUCGAUCCUUCUCGUCCAGCGUCGGUGGCAUUAGCGGAUUGAUGGAGGGUCGCACUUCGAUCUGCA----- .............((((((((((.(((..((((...(((((..(((.((..(((......))).)).)))...))))).......)).))..))).)))))))..))).......----- ( -39.40, z-score = -0.57, R) >droWil1.scaffold_180569 1268120 115 - 1405142 UCAAUGAGGAAAACAAUGCGGCCAUCUGGCAUGAGGGCCACAUCGCAAGUGCGAUCCUUUUCAUCCAAAGUGGGUGGCAUUAAUGGAUUAAUAGAAGGACGCACUUCGAUCUAUU----- .......(((......((.((((.((......)).)))).))(((.(((((((.((((((((((((.....)))))..(((((....)))))))))))))))))))))))))...----- ( -38.20, z-score = -2.69, R) >droVir3.scaffold_12823 907091 116 - 2474545 UCUAUGAGGAAUACAAUGCGUCCAUCCGGCAUCAAAGCCACAUCGCAGGUGCGAUCCUUUUCAUCCAACGUCGGCGGCAUUAGCGGAUUUAUCGAUGGACGCACCUCGAUCUGAUU---- ....(((((.......((((((((((.(((......)))...((((..((((((......))......((....))))))..)))).......)))))))))))))))........---- ( -40.01, z-score = -2.46, R) >droMoj3.scaffold_6496 26026823 116 - 26866924 UCGAUGAGAAAGACUAUGCGUCCGUCUGGCAUUAAAGCCACAUCGCAGGUGCGAUCCUUUUCAUCCAAUGUGGGUGGCAUUAAGGGAUUGAUAGAAGGACGCACUUCAAUCUAAGU---- .((((((((..(((.....)))..)))(((......))).))))).(((((((.((((((((((((.....)))))..(((((....)))))))))))))))))))..........---- ( -37.90, z-score = -2.32, R) >droGri2.scaffold_15245 5718481 112 + 18325388 UCUAUGAGGAAUACAAUGCGUCCGUCCGGCAUCAAGGCCACAUCGCAUGUGCGAUCCUUUUCAUCCAGCGUUGGCGGCAUCAACGGAUUUAUGGAGGGACGCACCUCGAUCU-------- ....(((((.......(((((((.((((..(((((((....(((((....))))))))).........((((((.....)))))))))...)))).))))))))))))....-------- ( -39.31, z-score = -1.71, R) >anoGam1.chr2L 9716315 120 + 48795086 UCAACCAUGACCACGAGUCGUCCAUCGGGCAGCAGCGCAACAUCGCUGGGUCGAUCUUUUUCGUCCAUACCUAUUGCUAGCAACCCAUUAGACGAAGGACGUAUUUCAACCUGUACCAAA ..............(((.(((((.(((((((((((((......)))))((.(((......))).)).......))))).......(....).))).)))))...)))............. ( -28.80, z-score = -0.80, R) >consensus UCAAUGAGGAACACAAUGCGACCGUCUGGCAUCAUGGCCACAUCGCAAGUGCGAUCCUUCUCGUCCAGCGUGGGCGGCAUCAACGGAUUGAUGGAGGGUCGCACCUCGAUCUAAAU____ ....(((((.......(((((((.((((((......))).............(((((...(((((.......))))).......)))))...))).))))))))))))............ (-25.31 = -25.21 + -0.10)
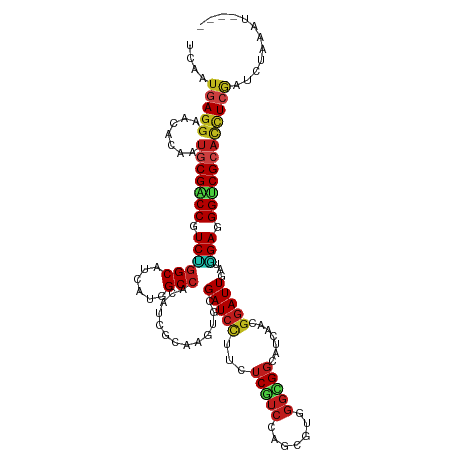
| Location | 3,370,999 – 3,371,106 |
|---|---|
| Length | 107 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 66.85 |
| Shannon entropy | 0.72075 |
| G+C content | 0.47110 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -10.13 |
| Energy contribution | -9.85 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
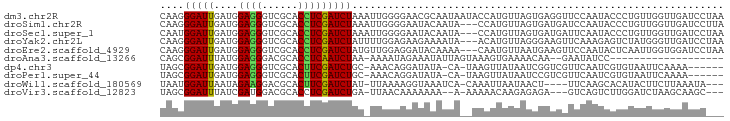
>dm3.chr2R 3370999 107 + 21146708 CAUCGCAAGUGCGAUCUUUCUCGUCCAGCGUGGGCGGCAUCAAGGGAUUGAUGGAGGGUCGCACCUCGAUCUAAAUUGGGGAACGCAAUAAU-ACCAUGUUAGUGAGG------ ..((((...((((((((((.((((((.....))))))((((((....))))))))))))))))(((((((....)))))))...........-.........))))..------ ( -37.00, z-score = -2.03, R) >droSim1.chr2R 2092388 104 + 19596830 CAUCGCAAGUGCGAUCUUUCUCGUCCAGCGUGGGCGGCAUCAAGGGAUUGAUGGAGGGUCGCACCUCGAUCUAAAUUGGGGAAUACAAUA----CCAUGUUAGUGAUG------ ((((((...((((((((((.((((((.....))))))((((((....))))))))))))))))(((((((....))))))).........----........))))))------ ( -39.60, z-score = -3.22, R) >droSec1.super_1 1028252 104 + 14215200 CAUCGCAAGUGCGAUCUUUCUCGUCCAGCGUGGGCGGCAUCAAUGGAUUGAUGGAGGGUCGCACCUCGAUCUAAAUUGGGGAAUACAAUA----CCAUGUUAGUGAUG------ ((((((...((((((((((.((((((.....))))))((((((....))))))))))))))))(((((((....))))))).........----........))))))------ ( -38.80, z-score = -2.99, R) >droYak2.chr2L 16049579 104 + 22324452 CAUCGCAAGUGCGAUCUUUCUCGUCCAGCGUGGGCGGCAUCAAGGGAUUGAUGGAGGGUCGCACCUCGAUCUAUUUUGGAGAAGAAAAUA----ACAUGUUAGGGAAG------ ..((.(..(((((((((((.((((((.....))))))((((((....)))))))))))))))))(((.(.......).))).........----........).))..------ ( -33.00, z-score = -1.69, R) >droEre2.scaffold_4929 7413638 104 - 26641161 CAUCGCAAGUGCGAUCUUUCUCGUCCAGCGUGGGCGGCAUCAAGGGAUUGAUGGAGGGUCGCACCUCGAUCUAUGUUGGAGGAUACAAAA----CAAUGUUAAUGAAG------ .(((((....))))).....((.((((((((((((..((((((....))))))((((......))))).))))))))))).)).......----..............------ ( -35.50, z-score = -2.37, R) >droAna3.scaffold_13266 3476720 95 - 19884421 CGUCGCAGGUGCGAUCCUUCUCGUCCAGGGUGGGUGGCAUCAGCGGAUUUAUGGAGGGACGCACCUCAAUCUAAAAAAUAGAAAUAUUAGUAAAG------------------- ....(.(((((((.(((((((.((((..((((.....))))...))))....))))))))))))))).........((((....)))).......------------------- ( -30.70, z-score = -2.75, R) >dp4.chr3 7405710 97 - 19779522 CAUCGCAGGUUCGAUCCUUCUCGUCCAGCGUCGGUGGCAUUAGCGGAUUGAUGGAGGGUCGCACUUCGAUCUGCAAACAGGAUAUACAUAAG--UUAUA--------------- ..(((.((((.(((((((((((((((.((.............))))).))).))))))))).)))))))((((....))))...........--.....--------------- ( -28.12, z-score = -1.01, R) >droPer1.super_44 58599 97 - 614636 CAUCGCAGGUUCGAUCCUUCUCGUCCAGCGUCGGUGGCAUUAGCGGAUUGAUGGAGGGUCGCACUUCGAUCUGCAAACAGGAUAUACAUAAG--UUAUA--------------- ..(((.((((.(((((((((((((((.((.............))))).))).))))))))).)))))))((((....))))...........--.....--------------- ( -28.12, z-score = -1.01, R) >droWil1.scaffold_180569 1268160 102 - 1405142 CAUCGCAAGUGCGAUCCUUUUCAUCCAAAGUGGGUGGCAUUAAUGGAUUAAUAGAAGGACGCACUUCGAUCUAUUUAAAAGGUAAAUCACAAAUUAAUAACU------------ .((((.(((((((.((((((((((((.....)))))..(((((....)))))))))))))))))))))))................................------------ ( -27.00, z-score = -3.55, R) >droVir3.scaffold_12823 907131 111 - 2474545 CAUCGCAGGUGCGAUCCUUUUCAUCCAACGUCGGCGGCAUUAGCGGAUUUAUCGAUGGACGCACCUCGAUCUGAUUAACAAAAAAAAAAAAACAAGAGAGAGUCAGUCUUG--- .((((.(((((((.(((...((((((..((....))((....)))))).....)).))))))))))))))((((((..(..................)..)))))).....--- ( -28.47, z-score = -1.30, R) >droMoj3.scaffold_6496 26026863 108 - 26866924 CAUCGCAGGUGCGAUCCUUUUCAUCCAAUGUGGGUGGCAUUAAGGGAUUGAUAGAAGGACGCACUUCAAUCUAAGUAAAAGAGGGAGAGAGAGAUGAUAUUAGUUAUG------ ((((.(.((((((.((((((((((((.....)))))..(((((....))))))))))))))))))((..(((.......)))..))....).))))............------ ( -28.80, z-score = -1.93, R) >droGri2.scaffold_15245 5718521 91 + 18325388 CAUCGCAUGUGCGAUCCUUUUCAUCCAGCGUUGGCGGCAUCAACGGAUUUAUGGAGGGACGCACCUCGAUCUAUACUCGAAAUUAGGAUUA----------------------- ..((((....))))((((((((......((((((.....)))))).((..((.((((......)))).))..))....))))..))))...----------------------- ( -22.90, z-score = -0.22, R) >anoGam1.chr2L 9716355 114 + 48795086 CAUCGCUGGGUCGAUCUUUUUCGUCCAUACCUAUUGCUAGCAACCCAUUAGACGAAGGACGUAUUUCAACCUGUACCAAAAUAACAAAAAAGGCAAAGAAAAACAUUACGAACA ..(((.((((.(((......)))))))..(((.(((((((.......)))).))))))..((.((((..(((..................)))....)))).))....)))... ( -15.57, z-score = 0.61, R) >consensus CAUCGCAAGUGCGAUCCUUCUCGUCCAGCGUGGGCGGCAUCAACGGAUUGAUGGAGGGUCGCACCUCGAUCUAAAUAAGAGAAAAAAAUAA___CCAUAUUA___A_G______ .((((......)))).....(((((.......))))).......(((((....((((......))))))))).......................................... (-10.13 = -9.85 + -0.28)
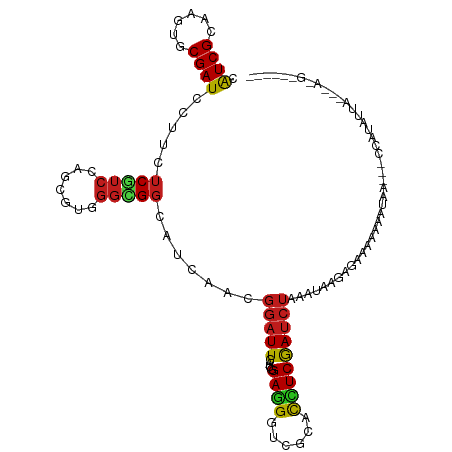
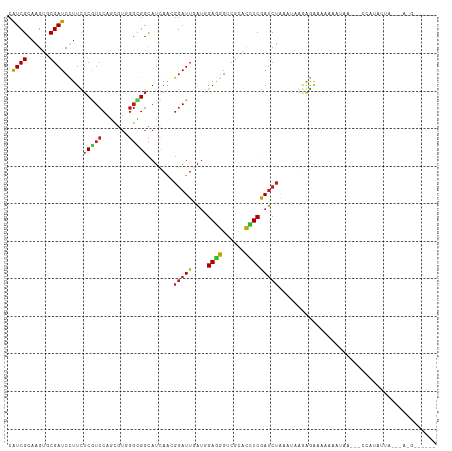
| Location | 3,371,039 – 3,371,133 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 58.78 |
| Shannon entropy | 0.88190 |
| G+C content | 0.39604 |
| Mean single sequence MFE | -18.63 |
| Consensus MFE | -7.01 |
| Energy contribution | -6.76 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.55 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3371039 94 + 21146708 CAAGGGAUUGAUGGAGGGUCGCACCUCGAUCUAAAUUGGGGAACGCAAUAAUACCAUGUUAGUGAGGUUCCAAUACCCUGUUGGUUGAUCCUAA ...(((((..(((.(((((.(.((((((..((((..(((..............)))..)))))))))).)....))))).)..))..))))).. ( -27.24, z-score = -0.74, R) >droSim1.chr2R 2092428 91 + 19596830 CAAGGGAUUGAUGGAGGGUCGCACCUCGAUCUAAAUUGGGGAAUACAAUA---CCAUGUUAGUGAUGAUCCAAUACCCUGUUGGUUGAUCCUUA ..((((((..(((((..(((((.(((((((....)))))))...(((...---...)))..)))))..))))....((....)))..)))))). ( -27.60, z-score = -1.17, R) >droSec1.super_1 1028292 91 + 14215200 CAAUGGAUUGAUGGAGGGUCGCACCUCGAUCUAAAUUGGGGAAUACAAUA---CCAUGUUAGUGAUGAUUCAAUACCCUGUUGGUUGAUCCUAA ....((((..(((((..(((((.(((((((....)))))))...(((...---...)))..)))))..))))....((....)))..))))... ( -22.70, z-score = -0.16, R) >droYak2.chr2L 16049619 91 + 22324452 CAAGGGAUUGAUGGAGGGUCGCACCUCGAUCUAUUUUGGAGAAGAAAAUA---ACAUGUUAGGGAAGUUCAAAGAGUCUAUGGGUUGAUCCUAA (((((....(((.((((......)))).)))..)))))............---.....((((((.((..((.((...)).))..))..)))))) ( -19.60, z-score = 0.09, R) >droEre2.scaffold_4929 7413678 91 - 26641161 CAAGGGAUUGAUGGAGGGUCGCACCUCGAUCUAUGUUGGAGGAUACAAAA---CAAUGUUAAUGAAGUUCCAAUACUCAAUUGGUGGAUCCUAA ...(((((((((.((((......)))).)))..((((((((...(((...---...)))........))))))))...........)))))).. ( -22.60, z-score = -0.61, R) >droAna3.scaffold_13266 3476760 72 - 19884421 CAGCGGAUUUAUGGAGGGACGCACCUCAAUCUAA-AAAAUAGAAAUAUUAGUAAAGUGAAAACAA--GAAUAUCC------------------- ....((((.....((((......))))..(((..-..((((....))))......((....)).)--))..))))------------------- ( -12.50, z-score = -2.03, R) >dp4.chr3 7405750 85 - 19779522 UAGCGGAUUGAUGGAGGGUCGCACUUCGAUCUGC-AAACAGGAUAUA-CA-UAAGUUAUAAUCGGUCGUUCAAUCGUGUAAUUCAAAA------ ..(((((((((.(..........).)))))))))-............-..-..(((((((..((((......))))))))))).....------ ( -15.50, z-score = 0.08, R) >droPer1.super_44 58639 85 - 614636 UAGCGGAUUGAUGGAGGGUCGCACUUCGAUCUGC-AAACAGGAUAUA-CA-UAAGUUAUAAUCCGUCGUUCAAUCGUGUAAUUCAAAA------ ..(((((((((..((((((((.....))))))..-.....((((..(-(.-...))....)))).))..)))))).))).........------ ( -16.50, z-score = -0.12, R) >droWil1.scaffold_180569 1268200 85 - 1405142 UAAUGGAUUAAUAGAAGGACGCACUUCGAUCUAU-UUAAAAGGUAAAUCA-CAAAUUAAUAACU----UUCAAGCACAUACUUCUUAAAUA--- ....(((((....((((......)))))))))((-((((.(((((.....-.............----..........))))).)))))).--- ( -8.29, z-score = -0.70, R) >droVir3.scaffold_12823 907171 84 - 2474545 UAGCGGAUUUAUCGAUGGACGCACCUCGAUCUGA-UUAACAAAAAAA--A-AAAAACAAGAGAGA---GUCAGUCUUGGAUCUAAGCAAGC--- ..(((((((((.....((((....(((..(((..-............--.-.......)))..))---)...)))))))))))..))....--- ( -13.75, z-score = -0.12, R) >consensus CAACGGAUUGAUGGAGGGUCGCACCUCGAUCUAA_UUAGAGAAUACAAUA__AAAAUGUUAGCGAUGGUUCAAUACUCUAUUGGUUGAUCC___ ....(((((....((((......))))))))).............................................................. ( -7.01 = -6.76 + -0.25)
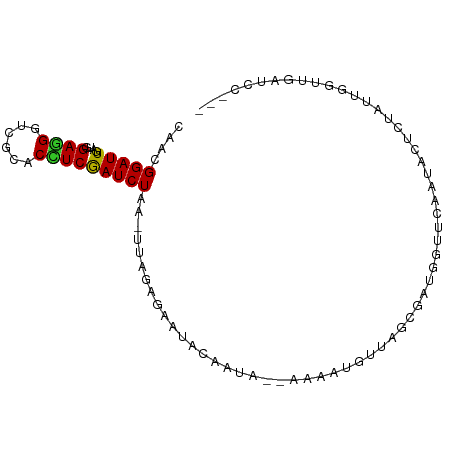
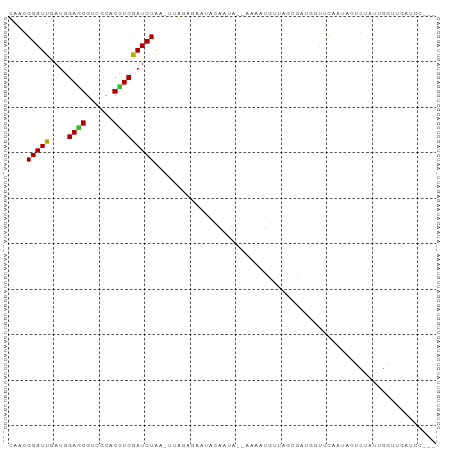
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:40 2011