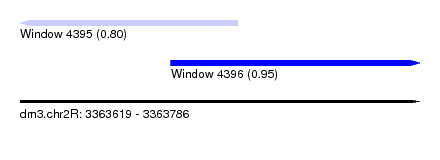
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,363,619 – 3,363,786 |
| Length | 167 |
| Max. P | 0.946327 |

| Location | 3,363,619 – 3,363,710 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 64.79 |
| Shannon entropy | 0.56437 |
| G+C content | 0.31013 |
| Mean single sequence MFE | -15.94 |
| Consensus MFE | -6.55 |
| Energy contribution | -5.55 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3363619 91 - 21146708 ---------UAUUUUAUUAUGUUUAUCCCCAAGUCAAAAUGUACAAAUAUUUAUGAAAAGGGAUCAAACACAGC---UGCACUUUCAAAUUAAUAAAGAAAUU ---------..((((((((((((((((((....(((((((((....)))))).)))...))))).)))))....---((......))...))))))))..... ( -14.40, z-score = -1.87, R) >droSec1.super_1 1020748 100 - 14215200 GUUAAUAAACAUUUAAGCACGCUUAUCCCCAAGUCAAAAUGUACAAAUAUUUAUGAAAAGGGGUCAAACACACG---UGCACUUUCAAAUUAAAAAAGAAACU ................(((((.....((((...(((((((((....)))))).)))...)))).........))---)))....................... ( -17.94, z-score = -2.73, R) >droSim1.chr2R 2084836 100 - 19596830 GUUAAUAAACAUUUAAGCACGCUUAUCCCCAAGUCAAAAUGUACAAAUAUUUAUGAAAAGGGAUCAAACACACC---UGCACUUUCAAAUUAAAAAAUAAACU (((..........((((....))))((((....(((((((((....)))))).)))...))))...))).....---.......................... ( -11.00, z-score = -1.07, R) >droPer1.super_44 51289 98 + 614636 ----AAUUAGGUUUGAAUCUGGAUCCGUGCUGAAAAAGCCGUUC-AAUGGUUAUAAUCGUGUCUCAGUUUCAGCUGUUGCACUUUUCCUUUUCAACAUAUACC ----..((((((....))))))....(((.((((((((((((..-.))))))......((((..((((....))))..))))......)))))).)))..... ( -20.40, z-score = -1.31, R) >consensus ____AUAAACAUUUAAGCACGCUUAUCCCCAAGUCAAAAUGUACAAAUAUUUAUGAAAAGGGAUCAAACACACC___UGCACUUUCAAAUUAAAAAAGAAACU ........................(((((....(((((((((....)))))).)))...)))))....................................... ( -6.55 = -5.55 + -1.00)


| Location | 3,363,682 – 3,363,786 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.80 |
| Shannon entropy | 0.19744 |
| G+C content | 0.28974 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -18.46 |
| Energy contribution | -20.13 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3363682 104 + 21146708 UUGACUUGGGGAUAAACAUAAUAAAAUAAUUG-----------CAAAUGAUCAAACAAAUCGUAUACAAUUCAAGCUUACGAUUUGAAGUCUUUAAAAGCUGAAACAACAAUAUA ..(((((...(((.....((((......))))-----------......)))...(((((((((..(.......)..))))))))))))))........................ ( -14.40, z-score = -1.32, R) >droSec1.super_1 1020811 115 + 14215200 UUGACUUGGGGAUAAGCGUGCUUAAAUGUUUAUUAACAUUUUCCAAAAGAUUAAACAAAUCGUAUACAAUUCAAGUUUGCGAUUUGAAGUCUUUAAAAGCUGAGACAACAAUAUA (((.((..(.(((((((((......)))))))))...........(((((((...(((((((((.((.......)).))))))))).))))))).....)..)).)))....... ( -26.80, z-score = -3.01, R) >droSim1.chr2R 2084899 115 + 19596830 UUGACUUGGGGAUAAGCGUGCUUAAAUGUUUAUUAACAUUUUGCAAAAGAUCAAACAAAUCGUAUACAAUUCAAGUUUGCGAUUUGAAGUCUUUAAAAGCUGAGACAACAAUAUA (((...((..(((((((((......)))))))))..)).....))).........(((((((((.((.......)).)))))))))..(((((........)))))......... ( -26.90, z-score = -2.65, R) >consensus UUGACUUGGGGAUAAGCGUGCUUAAAUGUUUAUUAACAUUUU_CAAAAGAUCAAACAAAUCGUAUACAAUUCAAGUUUGCGAUUUGAAGUCUUUAAAAGCUGAGACAACAAUAUA (((.((..(.(((((((((......)))))))))...........((((((....(((((((((.((.......)).)))))))))..)))))).....)..)).)))....... (-18.46 = -20.13 + 1.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:37 2011