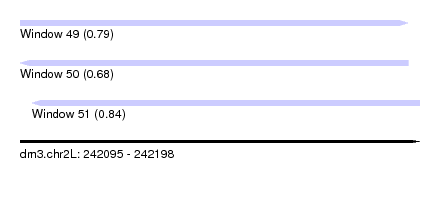
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 242,095 – 242,198 |
| Length | 103 |
| Max. P | 0.839518 |

| Location | 242,095 – 242,195 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 77.64 |
| Shannon entropy | 0.42568 |
| G+C content | 0.47371 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -18.64 |
| Energy contribution | -19.13 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
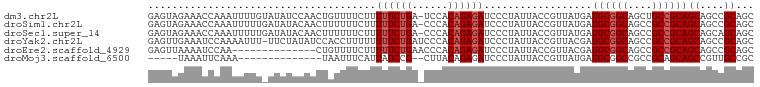
>dm3.chr2L 242095 100 + 23011544 GCUGCGGCUGCUGCGGCAGCUGCCGCAUCAUAACGGUAAUAGGGAUCUCUGUGGA-UCAGAAAAAGAAAACAGUUGGAUAUACAAAAUUUGGUUUCUACUC ((.((((((((....)))))))).)).............(((..(((((((....-.))))............(((......))).....)))..)))... ( -28.90, z-score = -1.75, R) >droSim1.chr2L 247300 100 + 22036055 GCUGCGGCUGCUGCGGCGGCUGCCGCAUCAUAACGGUAAUAGGGAUCUCUGUGGG-UCAGAAAAAGAAAAAAGUUGUAUAUCAAAAAUUUGGUUUCUACUC ((.((((((((....)))))))).)).............(((..(((((((....-.))))............(((.....)))......)))..)))... ( -27.10, z-score = -1.29, R) >droSec1.super_14 240041 100 + 2068291 GCUGCUGCUGCUGCGGCGGCUGCCGCAUCAUAACGGUAAUAGGGAUCUCUGUGGG-UCAGAAAAAGAAAAAAGUUGUAUAUCAAAAAUUUGGUUUCUACUC ((((((((....))))))))(((((........))))).(((..(((((((....-.))))............(((.....)))......)))..)))... ( -28.90, z-score = -2.06, R) >droYak2.chr2L 235107 100 + 22324452 GCUGCGGCUGCUGCGGCGGCUGCCGCAUCGUAACGGUAAUAGGGAUCUCUGUGGGAUCAGAAAAAAAAGGUGGAUAUAGAA-AAAUUUUGGAUUUCAACUC ((.((((((((....)))))))).))((((...))))......(((((.....))))).((((...(((((..........-..)))))...))))..... ( -25.90, z-score = -0.50, R) >droEre2.scaffold_4929 288368 87 + 26641161 GCUGCGGCUGCUGCGGCGGCUGCCGCAUCGUAACGGUAAUAGGGAUCUCUGUGGGUUCAGAAAAAGAAAACAG--------------UUGGAUUUUAACUC ((.((((((((....)))))))).))((((...))))....(((((((((((...(((.......))).))))--------------..)))))))..... ( -26.70, z-score = -1.02, R) >droMoj3.scaffold_6500 2472745 80 + 32352404 GCGGCAACGGCUGCUGCGGCGGCCGCAUCAUAACGGUAAUAGGGAUCUCUGUAAG--CGGAUAAUGAAAUUA--------------UUUGAAUUUA----- (((((..(.((....)).)..))))).((((..((.(.(((((....))))).).--))....)))).....--------------..........----- ( -22.90, z-score = -0.97, R) >consensus GCUGCGGCUGCUGCGGCGGCUGCCGCAUCAUAACGGUAAUAGGGAUCUCUGUGGG_UCAGAAAAAGAAAAAAGUUGUAUAU_AAAAAUUGGAUUUCUACUC ((.((((((((....)))))))).))...............((((((((((......)))).........(((((..........)))))))))))..... (-18.64 = -19.13 + 0.49)


| Location | 242,095 – 242,195 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 77.64 |
| Shannon entropy | 0.42568 |
| G+C content | 0.47371 |
| Mean single sequence MFE | -18.82 |
| Consensus MFE | -13.83 |
| Energy contribution | -14.33 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.678575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 242095 100 - 23011544 GAGUAGAAACCAAAUUUUGUAUAUCCAACUGUUUUCUUUUUCUGA-UCCACAGAGAUCCCUAUUACCGUUAUGAUGCGGCAGCUGCCGCAGCAGCCGCAGC .(((((.....((((.(((......)))..))))....((((((.-....))))))...)))))..........((((((.((((...)))).)))))).. ( -22.00, z-score = -1.11, R) >droSim1.chr2L 247300 100 - 22036055 GAGUAGAAACCAAAUUUUUGAUAUACAACUUUUUUCUUUUUCUGA-CCCACAGAGAUCCCUAUUACCGUUAUGAUGCGGCAGCCGCCGCAGCAGCCGCAGC .(((((....(((....)))..................((((((.-....))))))...)))))...((..((.((((((....)))))).))...))... ( -19.60, z-score = -1.40, R) >droSec1.super_14 240041 100 - 2068291 GAGUAGAAACCAAAUUUUUGAUAUACAACUUUUUUCUUUUUCUGA-CCCACAGAGAUCCCUAUUACCGUUAUGAUGCGGCAGCCGCCGCAGCAGCAGCAGC .(((((....(((....)))..................((((((.-....))))))...)))))...(((.((.((((((....)))))).))..)))... ( -20.70, z-score = -1.71, R) >droYak2.chr2L 235107 100 - 22324452 GAGUUGAAAUCCAAAAUUU-UUCUAUAUCCACCUUUUUUUUCUGAUCCCACAGAGAUCCCUAUUACCGUUACGAUGCGGCAGCCGCCGCAGCAGCCGCAGC (.((((.............-..................((((((......))))))..................((((((....)))))).)))))..... ( -18.70, z-score = -1.12, R) >droEre2.scaffold_4929 288368 87 - 26641161 GAGUUAAAAUCCAA--------------CUGUUUUCUUUUUCUGAACCCACAGAGAUCCCUAUUACCGUUACGAUGCGGCAGCCGCCGCAGCAGCCGCAGC ..............--------------((((......((((((......))))))................(.((((((....)))))).)....)))). ( -18.80, z-score = -0.99, R) >droMoj3.scaffold_6500 2472745 80 - 32352404 -----UAAAUUCAAA--------------UAAUUUCAUUAUCCG--CUUACAGAGAUCCCUAUUACCGUUAUGAUGCGGCCGCCGCAGCAGCCGUUGCCGC -----..........--------------.....((((....((--.....((......)).....))..)))).(((((.((.((....)).)).))))) ( -13.10, z-score = 0.46, R) >consensus GAGUAGAAACCAAAUUUUU_AUAUACAACUAUUUUCUUUUUCUGA_CCCACAGAGAUCCCUAUUACCGUUAUGAUGCGGCAGCCGCCGCAGCAGCCGCAGC ......................................((((((......))))))..................((((((....))))))((....))... (-13.83 = -14.33 + 0.50)

| Location | 242,098 – 242,198 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 76.96 |
| Shannon entropy | 0.43855 |
| G+C content | 0.47888 |
| Mean single sequence MFE | -20.81 |
| Consensus MFE | -15.13 |
| Energy contribution | -16.30 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
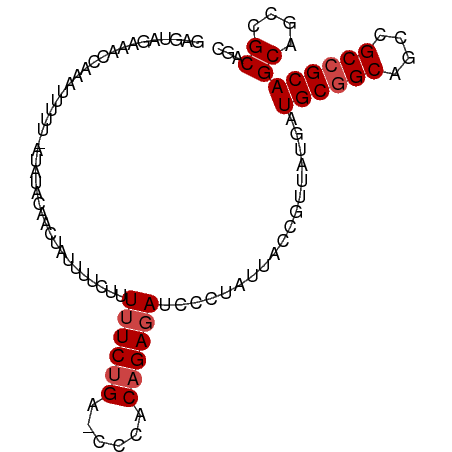
>dm3.chr2L 242098 100 - 23011544 GGCGAGUAGAAACCAAAUUUUGUAUAUCCAACUGUUUUCUUUUUCUGA-UCCACAGAGAUCCCUAUUACCGUUAUGAUGCGGCAGCUGCCGCAGCAGCCGC (((.(((((.....((((.(((......)))..))))....((((((.-....))))))...))))).......((.((((((....)))))).))))).. ( -24.10, z-score = -1.28, R) >droSim1.chr2L 247303 100 - 22036055 GGCGAGUAGAAACCAAAUUUUUGAUAUACAACUUUUUUCUUUUUCUGA-CCCACAGAGAUCCCUAUUACCGUUAUGAUGCGGCAGCCGCCGCAGCAGCCGC (((.(((((....(((....)))..................((((((.-....))))))...))))).......((.((((((....)))))).))))).. ( -24.00, z-score = -2.29, R) >droSec1.super_14 240044 100 - 2068291 GGCGAGUAGAAACCAAAUUUUUGAUAUACAACUUUUUUCUUUUUCUGA-CCCACAGAGAUCCCUAUUACCGUUAUGAUGCGGCAGCCGCCGCAGCAGCAGC ((..(((((....(((....)))..................((((((.-....))))))...))))).))(((.((.((((((....)))))).))..))) ( -21.00, z-score = -1.20, R) >droYak2.chr2L 235110 100 - 22324452 GGCGAGUUGAAAUCCAAAAUUU-UUCUAUAUCCACCUUUUUUUUCUGAUCCCACAGAGAUCCCUAUUACCGUUACGAUGCGGCAGCCGCCGCAGCAGCCGC .(((.((((.............-..................((((((......))))))..................((((((....)))))).))))))) ( -22.80, z-score = -1.77, R) >droEre2.scaffold_4929 288371 87 - 26641161 GGCGAGUUAAAAUCCAA--------------CUGUUUUCUUUUUCUGAACCCACAGAGAUCCCUAUUACCGUUACGAUGCGGCAGCCGCCGCAGCAGCCGC (((............((--------------(.((......((((((......))))))........)).)))..(.((((((....)))))).).))).. ( -22.84, z-score = -1.73, R) >droMoj3.scaffold_6500 2472748 77 - 32352404 --------UAAAUUCAA--------------AUAAUUUCAUUAUCCG--CUUACAGAGAUCCCUAUUACCGUUAUGAUGCGGCCGCCGCAGCAGCCGUUGC --------.........--------------......((((....((--.....((......)).....))..)))).(((((.((....)).)))))... ( -10.10, z-score = 1.06, R) >consensus GGCGAGUAGAAACCAAAUUUUU_AUAUACAACUAUUUUCUUUUUCUGA_CCCACAGAGAUCCCUAUUACCGUUAUGAUGCGGCAGCCGCCGCAGCAGCCGC (((......................................((((((......))))))..................((((((....))))))...))).. (-15.13 = -16.30 + 1.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:21 2011