| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,245,375 – 3,245,441 |
| Length | 66 |
| Max. P | 0.666178 |

| Location | 3,245,375 – 3,245,441 |
|---|---|
| Length | 66 |
| Sequences | 12 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 66.44 |
| Shannon entropy | 0.74888 |
| G+C content | 0.44679 |
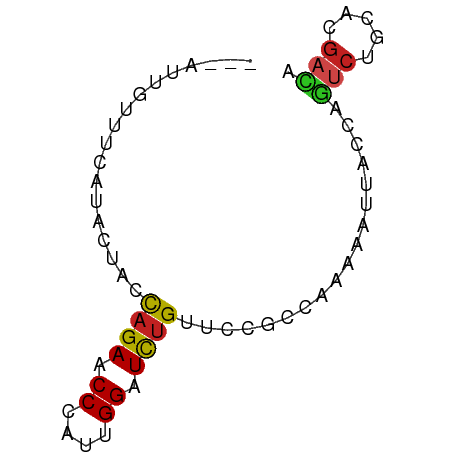
| Mean single sequence MFE | -8.82 |
| Consensus MFE | -5.54 |
| Energy contribution | -5.44 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.44 |
| Mean z-score | 0.15 |
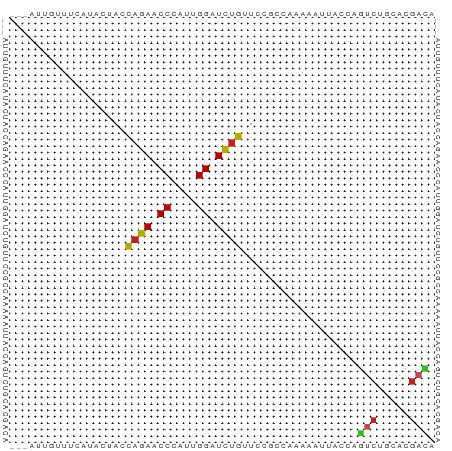
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
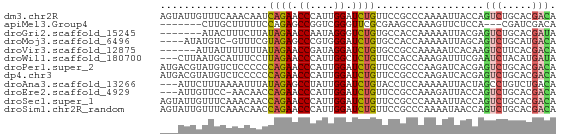
>dm3.chr2R 3245375 66 - 21146708 AGUAUUGUUUCAAACAAUCAGAACCCAUUGGAUCUGUUCCGCCCAAAAUUACCAGUCUGCACGACA ...(((((.....)))))((((.((....)).))))..................(((.....))). ( -7.80, z-score = 0.24, R) >apiMel3.Group4 9259960 56 + 10796202 -------CUUGCUUUUUCCAGAGCCGGUCGGGUUCGCGAAGCCAAAGUUCUCCA---CGAUCGACA -------...((((......))))((((((((...((.........))...)).---))))))... ( -13.40, z-score = 0.14, R) >droGri2.scaffold_15245 3860885 59 - 18325388 -------AUACUUUCUUAUAGAACCAAUAGGGUCUGUGCCACCAAAAAUUACGAGUCUGCACGAUA -------.........((((((.((....)).)))))).............((.(....).))... ( -6.40, z-score = 0.72, R) >droMoj3.scaffold_6496 10667828 61 + 26866924 ----AUAUGUC-GUUUCGUAGAGCCCGUGGGAUCUGUGCCACCAAAAAUUAGCAGUCUGCAUGACA ----...((((-((...(((((.((....))..((((..............))))))))))))))) ( -12.74, z-score = -0.03, R) >droVir3.scaffold_12875 10753083 60 - 20611582 ------AUUAUUUUUUUAUAGAACCGAUAGGAUCUGUGCCGCCAAAAAUCACAAGUCUUCACGACA ------..........((((((.((....)).))))))................(((.....))). ( -9.20, z-score = -1.24, R) >droWil1.scaffold_180700 337869 63 + 6630534 ---CUUAAUGCAUUUCCUUAGAACCCAUUGGCUCUGUUCCACCAAAGAUUUCGAAUCUACAUGAUA ---...............((((.((....)).)))).........((((.....))))........ ( -4.00, z-score = 1.11, R) >droPer1.super_2 1686334 66 - 9036312 AUGACGUAUGUCUCCCCCCAGAACCCAUUGGAUCUGUUCCGCCCAAGAUCACGAGUCUGCACGACA ........((((......((((.....((((((((..........))))).))).))))...)))) ( -11.10, z-score = 0.19, R) >dp4.chr3 1513680 66 - 19779522 AUGACGUAUGUCUCCCCCCAGAACCCAUUGGAUCUGUUCCGCCCAAGAUCACGAGUCUGCACGACA ........((((......((((.....((((((((..........))))).))).))))...)))) ( -11.10, z-score = 0.19, R) >droAna3.scaffold_13266 3453610 63 - 19884421 ---AUUCUUUAAAAUUUAUAGAGCCUAUUGGAUCUGUACCUCCAAAAAUUACUAGCCUGUCUGACA ---.............((((((.((....)).))))))............................ ( -7.00, z-score = -0.31, R) >droEre2.scaffold_4929 7292044 62 + 26641161 ---AUUGUUCC-AACAACCAGAACCCAUUGGAUCUGUUCCGCCAAAGAUUACCAGUCUGCACGACA ---..((((..-......((((.((....)).))))....((...((((.....))))))..)))) ( -8.10, z-score = 0.06, R) >droSec1.super_1 888614 66 - 14215200 AGUAUUGUUUCAAACAACCAGAACCCAUUGGAUCUGUUCCGCCCAAAAUUACCAGUCUGCACGACA .(((((((.....)))).((((.((....)).)))).............)))..(((.....))). ( -7.60, z-score = 0.32, R) >droSim1.chr2R_random 744239 66 - 2996586 AGUAUUGUUUCAAACAACCAGAACCCAUUGGAUCUGUUCCGCCCAAAAUAACCAGUCUGCACGACA ....((((((........((((.((....)).)))).........))))))...(((.....))). ( -7.43, z-score = 0.36, R) >consensus ___AUUGUUUCAUACUACCAGAACCCAUUGGAUCUGUUCCGCCAAAAAUUACCAGUCUGCACGACA ..................((((.((....)).))))..................(((.....))). ( -5.54 = -5.44 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:26 2011