| Sequence ID | dm3.chr2L |
|---|---|
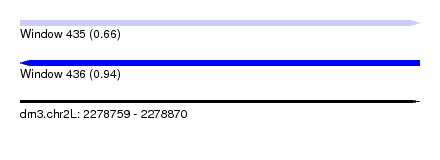
| Location | 2,278,759 – 2,278,870 |
| Length | 111 |
| Max. P | 0.941184 |

| Location | 2,278,759 – 2,278,870 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 65.47 |
| Shannon entropy | 0.66888 |
| G+C content | 0.52225 |
| Mean single sequence MFE | -28.09 |
| Consensus MFE | -9.64 |
| Energy contribution | -9.44 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.663567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
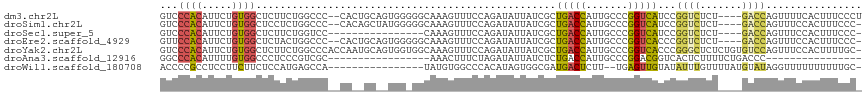
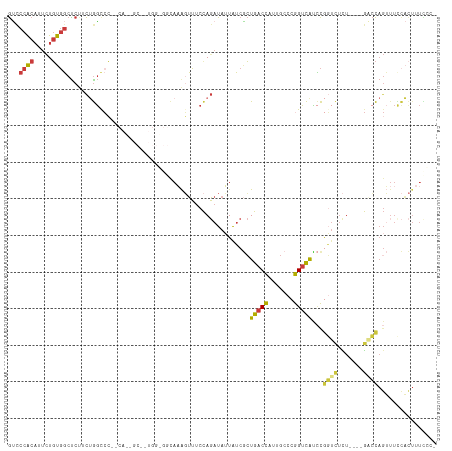
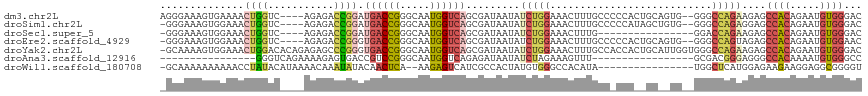
>dm3.chr2L 2278759 111 + 23011544 GUCCCACAUUCUGUGGCUCUUCUGGCCC--CACUGCAGUGGGGGCAAAGUUUCCAGAUAUUAUCGCUGACCAUUGCCCGGUCAUCCGGUCUCU----GACCAGUUUUCACUUUCCCU .........((((.((..(((.((.(((--(((....)))))).)))))..)))))).......((((..((..(.((((....)))).)..)----)..))))............. ( -33.10, z-score = -1.27, R) >droSim1.chr2L 2237401 110 + 22036055 GUCCCACAUUCUGUGGCUCCUCUGGCCC--CACAGCUAUGGGGGCAAAGUUUCCAGAUAUUAUCGCUGACCAUUGCCCGGUCAUCCGGUCUCU----GACCAGUUUCCACUUUCCC- (((((.(((.((((((..((...))..)--)))))..))))))))(((((.....(((...)))((((..((..(.((((....)))).)..)----)..))))....)))))...- ( -33.20, z-score = -1.60, R) >droSec1.super_5 453641 96 + 5866729 GUCCCACAUUCUGUGGCUCUUCUGGUCC----------------CAAAGUUUCCAGAUAUUAUCGCUGACCAUUGCCCGGUCAUCCGGUCUCU----GACCAGUUUCCACUUUCCC- ............((((.....((((((.----------------...((...((.(((...)))..(((((.......)))))...))...))----))))))...))))......- ( -21.40, z-score = -1.51, R) >droEre2.scaffold_4929 2315850 110 + 26641161 GUUCCACAUUCUGUGGCUCUACUGGCCC--CACUGCAGUGGGGGCAAAGUUUCCAGAUAUUAUCGCUGACCAUUGCCCGGUCACCCGGUCUCU----GACCAGUUUCCACUUUCCC- ............((((....((((((((--(((....))))))..........((((....((((.(((((.......)))))..)))).)))----).)))))..))))......- ( -34.50, z-score = -1.64, R) >droYak2.chr2L 2260191 116 + 22324452 GUCCCACAUUCUGUGGCUCUUCUGGCCCACCAAUGCAGUGGUGGCAAAGUUUCCAGAUAUUAUCGCUGACCAUUGCCCGGUCACCCGGGCUCUCUGUGUCCAGUUUCCACUUUUGC- (..((((.....))))..).(((((.((((((......))))))........))))).......((((..(((.((((((....)))))).....)))..))))............- ( -35.40, z-score = -1.47, R) >droAna3.scaffold_12916 9993687 84 - 16180835 GGCCCACAUUUUGUGGCCCUCCCGUCGC-----------------AAACUUUCUAGAUAUUAUCUCUGACCAUUGCCCGGACGGUCACUCUUUUCUGACCC---------------- ((.((((.....)))).))..(((..((-----------------((....((.((((...))))..))...)))).)))..(((((........))))).---------------- ( -20.80, z-score = -2.48, R) >droWil1.scaffold_180708 2242109 98 + 12563649 ACCCCGCCUCCUUCUUCUCCAUGAGCCA----------------UAUGUGGCCCACAUAGUGGCGAUGACUCUU--UGAGUUGUAUAUUUGUUUUAUGUAUAGGUUUUUUUUUUGC- .........(((.......(((((((((----------------((((((...))))).))))).(..((((..--.))))..).........)))))...)))............- ( -18.20, z-score = -0.62, R) >consensus GUCCCACAUUCUGUGGCUCUUCUGGCCC__CA__GC__UGG_GGCAAAGUUUCCAGAUAUUAUCGCUGACCAUUGCCCGGUCAUCCGGUCUCU____GACCAGUUUCCACUUUCCC_ ...((((.....))))..................................................(((((.......)))))...((((.......))))................ ( -9.64 = -9.44 + -0.20)
| Location | 2,278,759 – 2,278,870 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 65.47 |
| Shannon entropy | 0.66888 |
| G+C content | 0.52225 |
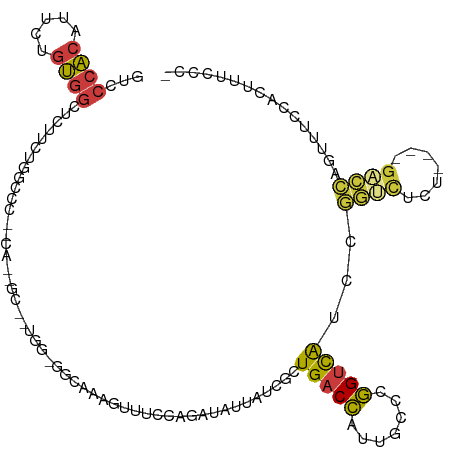
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -10.89 |
| Energy contribution | -10.91 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
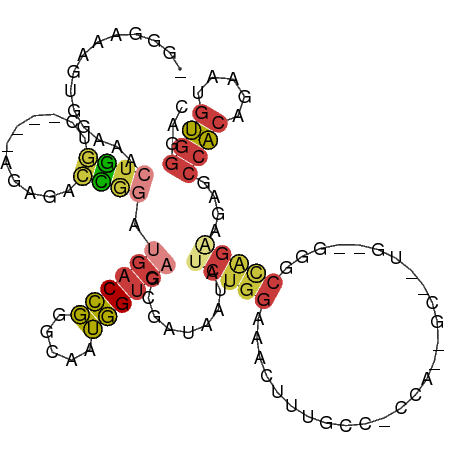
>dm3.chr2L 2278759 111 - 23011544 AGGGAAAGUGAAAACUGGUC----AGAGACCGGAUGACCGGGCAAUGGUCAGCGAUAAUAUCUGGAAACUUUGCCCCCACUGCAGUG--GGGCCAGAAGAGCCACAGAAUGUGGGAC ..............(((..(----(....((((....))))....))..)))........((((....)((((.((((((....)))--))).))))))).((((.....))))... ( -36.60, z-score = -1.39, R) >droSim1.chr2L 2237401 110 - 22036055 -GGGAAAGUGGAAACUGGUC----AGAGACCGGAUGACCGGGCAAUGGUCAGCGAUAAUAUCUGGAAACUUUGCCCCCAUAGCUGUG--GGGCCAGAGGAGCCACAGAAUGUGGGAC -((.((((((((..((((((----...)))))).((((((.....)))))).........)))....))))).))((((((.(((((--(..((...))..))))))..)))))).. ( -41.30, z-score = -2.47, R) >droSec1.super_5 453641 96 - 5866729 -GGGAAAGUGGAAACUGGUC----AGAGACCGGAUGACCGGGCAAUGGUCAGCGAUAAUAUCUGGAAACUUUG----------------GGACCAGAAGAGCCACAGAAUGUGGGAC -......((((...((((((----.(((.(((((((((((.....)))))).........)))))...)))..----------------.)))))).....))))............ ( -27.90, z-score = -1.81, R) >droEre2.scaffold_4929 2315850 110 - 26641161 -GGGAAAGUGGAAACUGGUC----AGAGACCGGGUGACCGGGCAAUGGUCAGCGAUAAUAUCUGGAAACUUUGCCCCCACUGCAGUG--GGGCCAGUAGAGCCACAGAAUGUGGAAC -......((((...(((..(----(....((((....))))....))..)))................((((((((((((....)))--)))...))))))))))............ ( -41.00, z-score = -2.14, R) >droYak2.chr2L 2260191 116 - 22324452 -GCAAAAGUGGAAACUGGACACAGAGAGCCCGGGUGACCGGGCAAUGGUCAGCGAUAAUAUCUGGAAACUUUGCCACCACUGCAUUGGUGGGCCAGAAGAGCCACAGAAUGUGGGAC -((...(((....))).(((.((....((((((....))))))..))))).)).......(((((........((((((......)))))).)))))....((((.....))))... ( -44.60, z-score = -3.21, R) >droAna3.scaffold_12916 9993687 84 + 16180835 ----------------GGGUCAGAAAAGAGUGACCGUCCGGGCAAUGGUCAGAGAUAAUAUCUAGAAAGUUU-----------------GCGACGGGAGGGCCACAAAAUGUGGGCC ----------------.(((((........))))).((((.((((...(((((.......))).))....))-----------------))..)))).((.((((.....)))).)) ( -27.10, z-score = -2.63, R) >droWil1.scaffold_180708 2242109 98 - 12563649 -GCAAAAAAAAAACCUAUACAUAAAACAAAUAUACAACUCA--AAGAGUCAUCGCCACUAUGUGGGCCACAUA----------------UGGCUCAUGGAGAAGAAGGAGGCGGGGU -...........((((....................((((.--..))))...((((.((.(((((((((....----------------)))))))))........)).)))))))) ( -20.90, z-score = -1.60, R) >consensus _GGGAAAGUGGAAACUGGUC____AGAGACCGGAUGACCGGGCAAUGGUCAGCGAUAAUAUCUGGAAACUUUGCC_CCA__GC__UG__GGGCCAGAAGAGCCACAGAAUGUGGGAC .............................(((((((((((.....)))))).........)))))....................................((((.....))))... (-10.89 = -10.91 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:42 2011