
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,208,343 – 3,208,463 |
| Length | 120 |
| Max. P | 0.893215 |

| Location | 3,208,343 – 3,208,463 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 66.96 |
| Shannon entropy | 0.56357 |
| G+C content | 0.46181 |
| Mean single sequence MFE | -30.25 |
| Consensus MFE | -12.76 |
| Energy contribution | -14.60 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.762615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3208343 120 + 21146708 ACCAAA-GCAUUGCAAGAGU-CGAGAUGGGAUCUUUCGGGCGAUCUUUGAAGUAUUUUCCAAGUAUCUCUAUCUCUAUCUCUAUCUCCGGCCAGAUCCCAUCUCACCCCAUUGGCCGUGUCU ......-((((.((..(.((-.((((((((((((.(((((.(((....((..((((.....))))..)).............))))))))..)))))))))))))).).....)).)))).. ( -31.43, z-score = -1.12, R) >droAna3.scaffold_13266 3419188 101 + 19884421 UCCAAA-GCAUAACAAGAAAACGAGAGUAGAUCUCU--GGUGGUUUUGGAAGAA--UUCUAAAAAGGAAA----------------CCACUUCGAUCACAUCUCGACGUGGCGACCGUGACA ((((((-((...........((.((((.....))))--.)).))))))))....--.........((...----------------((((.((((.......)))).))))...))...... ( -25.80, z-score = -1.25, R) >droEre2.scaffold_4929 7256037 98 - 26641161 ACCAAA-GCAUUGCAAGAGA-CGAGAUGGGAUCUUUGGGCUGUUUUUUAAAGU----CUCAAAGAUCU------------------CUAGCCAGAUCCGAUCUCGCCCCAUUGGCCGUGUCU ......-........(((.(-((((((.((((((...(((((.(((((.....----...)))))...------------------.))))))))))).)))))(((.....))).)).))) ( -30.10, z-score = -1.53, R) >droYak2.chr2L 15892958 107 + 22324452 ACCAAA-GCAUUGCAAGAGA-CGAGACAGGAUCUUUCGGGUGUUUUUCAAGAU---UUCUAAGUAUCUAAGUAU----------CCCCAACCAGAUCCAAUCUCGCCCCAUUGGCCGUGUCU ......-........(((.(-(((((..((((((...(((..(.(((..((((---........))))))).).----------.)))....))))))..))))(((.....))).)).))) ( -26.20, z-score = -1.52, R) >droSec1.super_1 851898 115 + 14215200 ACCAAAAGCAUUGCAAGGGU-CGAGAUGGGAUCUUUAGGAGGAUCUUUGAAGUAUUUUCCAAGUAUCUGUAUCUUUAU------CUCCGACCUGAACCCAUCUCGCCCCAUUGGCCGUGUCU .......((((.((..(((.-(((((((((...((((((.(((....((((((((.............))).))))).------.)))..))))))))))))))).)))....)).)))).. ( -37.72, z-score = -2.73, R) >consensus ACCAAA_GCAUUGCAAGAGA_CGAGAUGGGAUCUUUCGGGUGAUUUUUGAAGUA__UUCCAAGUAUCUAA_U_U__________C_CCAACCAGAUCCCAUCUCGCCCCAUUGGCCGUGUCU .......((((.((.......((((((.((((((...((.((.............................................)).)))))))).))))))........)).)))).. (-12.76 = -14.60 + 1.84)


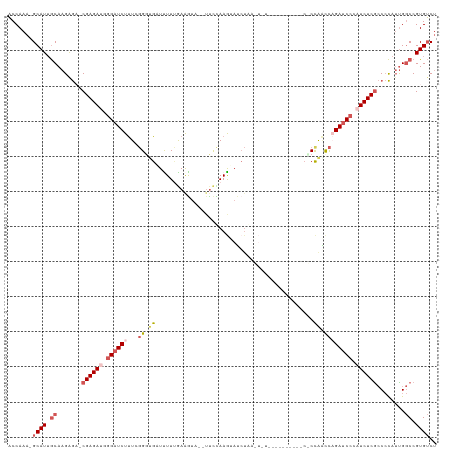
| Location | 3,208,343 – 3,208,463 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 66.96 |
| Shannon entropy | 0.56357 |
| G+C content | 0.46181 |
| Mean single sequence MFE | -33.61 |
| Consensus MFE | -13.67 |
| Energy contribution | -14.15 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3208343 120 - 21146708 AGACACGGCCAAUGGGGUGAGAUGGGAUCUGGCCGGAGAUAGAGAUAGAGAUAGAGAUACUUGGAAAAUACUUCAAAGAUCGCCCGAAAGAUCCCAUCUCG-ACUCUUGCAAUGC-UUUGGU ...((.(((....(((((((((((((((((...(((...((....))........(((..(((((......)))))..)))..)))..)))))))))))).-)))))......))-).)).. ( -37.80, z-score = -1.86, R) >droAna3.scaffold_13266 3419188 101 - 19884421 UGUCACGGUCGCCACGUCGAGAUGUGAUCGAAGUGG----------------UUUCCUUUUUAGAA--UUCUUCCAAAACCACC--AGAGAUCUACUCUCGUUUUCUUGUUAUGC-UUUGGA ......((..(((((.((((.......)))).))))----------------)..)).........--....((((((......--.((((...((....))..)))).......-)))))) ( -22.74, z-score = -0.15, R) >droEre2.scaffold_4929 7256037 98 + 26641161 AGACACGGCCAAUGGGGCGAGAUCGGAUCUGGCUAG------------------AGAUCUUUGAG----ACUUUAAAAAACAGCCCAAAGAUCCCAUCUCG-UCUCUUGCAAUGC-UUUGGU ...((.((((((.((((((((((.((((((((((((------------------((.((.....)----))))).......))))...)))))).))))))-)))))))....))-).)).. ( -34.01, z-score = -2.54, R) >droYak2.chr2L 15892958 107 - 22324452 AGACACGGCCAAUGGGGCGAGAUUGGAUCUGGUUGGGG----------AUACUUAGAUACUUAGAA---AUCUUGAAAAACACCCGAAAGAUCCUGUCUCG-UCUCUUGCAAUGC-UUUGGU ...((.((((((.((((((((((.((((((..(((((.----------......((((........---)))).........))))).)))))).))))))-)))))))....))-).)).. ( -38.89, z-score = -3.65, R) >droSec1.super_1 851898 115 - 14215200 AGACACGGCCAAUGGGGCGAGAUGGGUUCAGGUCGGAG------AUAAAGAUACAGAUACUUGGAAAAUACUUCAAAGAUCCUCCUAAAGAUCCCAUCUCG-ACCCUUGCAAUGCUUUUGGU .......(((((.(((((((((((((.((.....((((------...........(((..(((((......)))))..)))))))....)).)))))))))-.))))..........))))) ( -34.60, z-score = -1.47, R) >consensus AGACACGGCCAAUGGGGCGAGAUGGGAUCUGGCUGG_G__________A_A_AUAGAUACUUGGAA__UACUUCAAAAAACACCCGAAAGAUCCCAUCUCG_UCUCUUGCAAUGC_UUUGGU ...((..(.(((.(((.((((((.(((((............................................................))))).)))))).))).))))..))........ (-13.67 = -14.15 + 0.48)


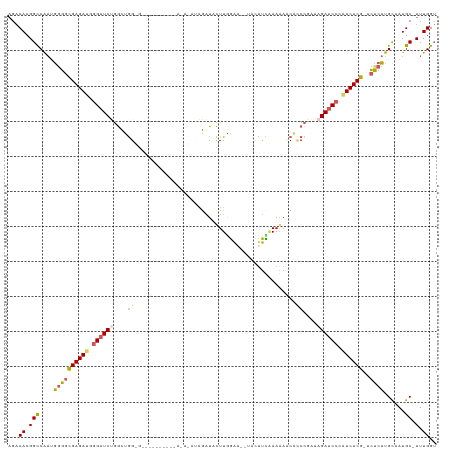
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:25 2011