
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,195,443 – 3,195,542 |
| Length | 99 |
| Max. P | 0.542445 |

| Location | 3,195,443 – 3,195,542 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.98 |
| Shannon entropy | 0.50919 |
| G+C content | 0.45914 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -11.48 |
| Energy contribution | -10.06 |
| Covariance contribution | -1.43 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3195443 99 + 21146708 --GCAACAGCGUC-GCGCCCAAUGAGGCGUAUGCGCAAUAAAAUUAAGCAUACGC-UGCCCGCUGAUGGGAAAUGCAUUCG------ACUAACAAGAAAUGCAUUAAAA-- --.((.(((((..-(((((......)))((((((.............))))))..-.)).))))).))...(((((((((.------........).))))))))....-- ( -29.42, z-score = -1.51, R) >droPer1.super_2 1635948 108 + 9036312 --GCAACAGAGCC-GCACUCAAUGGGGCGUAUUUGCCAUAAAGUUUGGUAUACGCUCAGCGGUGUCUGGGAAAUGCAUUUGCAUUUGAUUGUCAAGAAGUGCAAUUGACAA --....(((((((-((.(.....)((((((((..((((.......)))))))))))).))))).))))..((((((....))))))..(((((((.........))))))) ( -37.60, z-score = -2.55, R) >dp4.chr3 1463490 108 + 19779522 --GCAACAGAGCC-GCACUCAAUGGGGCGUAAUUGCCAUAAAGUUAGGUAUACGCUCAGCGGUGUCUGGGAAAUGCAUUUGCAUUUGAUUGUCAAGAAGUGCAAUUGACAA --....(((((((-((.(.....)(((((((..((((.........))))))))))).))))).))))..((((((....))))))..(((((((.........))))))) ( -35.30, z-score = -2.14, R) >droAna3.scaffold_13266 3406172 93 + 19884421 --GCAACAGCCUCUCCGCUCAAUGAGGCGUAUGUGCAAUAAG-UUAAGUAUACGC-CACCCAUGGCUGGGAAAUGCAUUUU------AUUAGCUGGAAAUUCC-------- --((....))...((((((.(((((((((((((.((.....)-)....)))))))-).((((....)))).........))------)))))).)))......-------- ( -24.00, z-score = -0.57, R) >droEre2.scaffold_4929 7242097 99 - 26641161 --GCAACAGCGCG-CCGCUCAAUGAGGCGUAUGCGCAAUAAAAUUAAGUAUACGC-UGCCCGGGGGUGGGAAGUGCAUUCG------GCUAACAAGAAAUGUAAUAAAA-- --((....((((.-((((((.....(((((((((.............))))))))-)......))))))...)))).....------))....................-- ( -29.12, z-score = -0.48, R) >droYak2.chr2L 15879622 99 + 22324452 --GCAACAGCGUC-CCGCUCAAUGAGGCGUAUGCGCAAUAAAAUUAAGUAUACGC-UGCUCGGGGAUGGGAAAUGCAUUCG------GCUAACAAGAAAUGUAAUAAAA-- --((....)).((-((((((.....(((((((((.............))))))))-).....))).)))))..((((((..------.((....)).))))))......-- ( -27.52, z-score = -1.51, R) >droSec1.super_1 839049 99 + 14215200 --GCAACAGCGUC-GCGUCCAAUGAGGCGUAUGCGCAAUAAAAUUAAGCAUACGC-CGCCCGCGGAUGGGAAAUGCAUUCG------ACUAACAAGAAAUGCAUUAAAA-- --((....)).((-.(((((.....(((((((((.............))))))))-)(....)))))).))(((((((((.------........).))))))))....-- ( -33.32, z-score = -2.61, R) >droSim1.chr2R_random 674478 99 + 2996586 --GCAACAGCGUC-GCGUCCAAUGAGGCGUAUGCGCAAUAAAAUUAAGCAUACGC-CGCCCGCGGAUGGGAAAUGCAUUCG------ACUAACAAGAAAUGCAUUAAAA-- --((....)).((-.(((((.....(((((((((.............))))))))-)(....)))))).))(((((((((.------........).))))))))....-- ( -33.32, z-score = -2.61, R) >droWil1.scaffold_180700 2841273 105 + 6630534 AGGCAACAACAGC-GAGCGCAAUGAGGCGUAUGCGCAAUAAAAUUAAGUAUACGCUAUGCCUUCUAUGGGAAAUGUAUGAACA---GAUACACACUCAUUGCGAUUGAA-- .(....).....(-((.(((((((((((((((((.............))))))))....(((.....)))...(((((.....---.)))))..))))))))).)))..-- ( -32.12, z-score = -3.12, R) >consensus __GCAACAGCGUC_GCGCUCAAUGAGGCGUAUGCGCAAUAAAAUUAAGUAUACGC_CGCCCGUGGAUGGGAAAUGCAUUCG______ACUAACAAGAAAUGCAAUAAAA__ .................((((.....((((((((.............))))))))...(.....).))))...(((((((...............))).))))........ (-11.48 = -10.06 + -1.43)
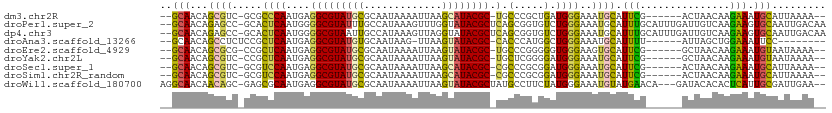

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:23 2011