
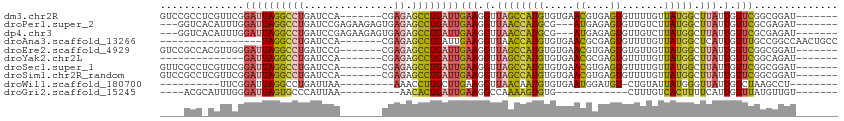
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,195,315 – 3,195,414 |
| Length | 99 |
| Max. P | 0.924394 |

| Location | 3,195,315 – 3,195,414 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 68.96 |
| Shannon entropy | 0.60623 |
| G+C content | 0.46780 |
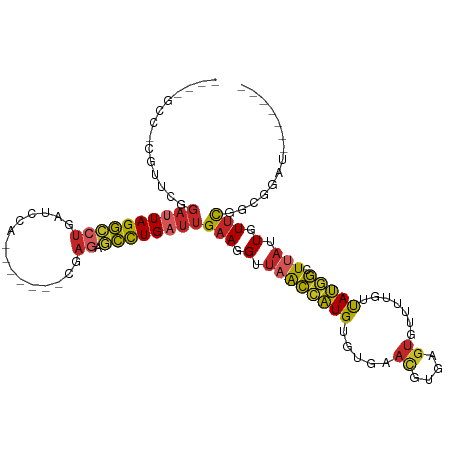
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -11.26 |
| Energy contribution | -12.63 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3195315 99 + 21146708 GUCCGCCUCGUUCGGAUUAGGCCUGAUCCA-------CGAGAGCCUGAUUGAAGGUUAGCCAUGUGUGAACGUGAGUGUUUUGUUAUGGCUUAUUGUUCGGCGGAU------- (((((((.......((((((((((......-------..)).))))))))(((.(..(((((((...(((((....)))))...)))))))...).))))))))))------- ( -38.10, z-score = -3.50, R) >droPer1.super_2 1635815 100 + 9036312 ---GGUCACAUUUGGAUUAGGCCUGAUCCGAGAAGAGUGAGAGCCUGAUUGAAGGUUAACCAUGCG---AUGAGAGUGUUGUCUUAUGGCUUAUUGUUCGCGAGAU------- ---.(((....((((((((....))))))))(((.(((((((((((......)))))..(((((.(---(..(.....)..)).))))))))))).)))....)))------- ( -29.50, z-score = -1.81, R) >dp4.chr3 1463357 100 + 19779522 ---GGUCACAUUUGGAUUAGGCCUGAUCCGAGAAGAGUGAGAGCCUGAUUGAAGGUUAACCAUGCG---AUGAGAGUGUUGUCUUAUGGCUUAUUGUUCGCGAGAU------- ---.(((....((((((((....))))))))(((.(((((((((((......)))))..(((((.(---(..(.....)..)).))))))))))).)))....)))------- ( -29.50, z-score = -1.81, R) >droAna3.scaffold_13266 3406045 89 + 19884421 -----------------UAGGCCUGAUCCA-------CGAGAGCCUGAUUGAAGGUUAACCAUGUGUGAACGCGAGUGUUUUGUUAUGGCUCAUUGUUUGCCGGCCAACUGCC -----------------..((((.(....(-------((((((((((((.(((((....)).((((....))))....))).)))).))))).))))...).))))....... ( -27.20, z-score = -1.64, R) >droEre2.scaffold_4929 7241969 99 - 26641161 GUCCGCCACGUUGGGAUUAGGCCUGAUCCG-------CGAGAGCCUGAUUGAAGGUUAGCCAUGUGUGAACGUGAGUGUGUUGUUAUGGCUUAUUGUUCGGCGGAU------- (((((((.......((((((((((......-------..)).))))))))(((.(..(((((((.....((....)).......)))))))...).))))))))))------- ( -38.50, z-score = -3.27, R) >droYak2.chr2L 15879508 85 + 22324452 --------------GAUUAGGCCUGAUCCA-------CGAGAGCCUGAUUGAAGGUUAGCCAUGUGUGAACGCGAGUGUUUUGUUAUGGCUUAUUGUUCGGCAGAU------- --------------((((((((((......-------..)).))))))))(((.(..(((((((...(((((....)))))...)))))))...).))).......------- ( -26.20, z-score = -2.06, R) >droSec1.super_1 838921 99 + 14215200 GUUCGCCUCGUUCGGAUUAGGCCUGAUCCA-------CGAGAGCCUGAUUGAAGGUUAGCCAUGUGUGAACGUGAGUGUUUUGUUAUGGCUUAUUGUUCGGCGGAU------- (((((((.......((((((((((......-------..)).))))))))(((.(..(((((((...(((((....)))))...)))))))...).))))))))))------- ( -35.20, z-score = -2.71, R) >droSim1.chr2R_random 674350 99 + 2996586 GUCCGCCUCGUUCGGAUUAGGCCUGAUCCA-------CGAGAGCCUGAUUGAAGGUUAGCCAUGUGUGAACGUGAGUGUUUUGUUAUGGCUUAUUGUUCGGCGGAU------- (((((((.......((((((((((......-------..)).))))))))(((.(..(((((((...(((((....)))))...)))))))...).))))))))))------- ( -38.10, z-score = -3.50, R) >droWil1.scaffold_180700 2841155 85 + 6630534 ----------UUCGGAUUAGGCCUGAUUAA---------AAACCUUGCUUGAAGGUUAACAAUGUGUGAAUGGAUGU-CUGUAUUAUGGGUUAUUGUCUAAGCCU-------- ----------...((.((((((........---------.((((((.....))))))...(((.(((((((((....-)))).))))).)))...)))))).)).-------- ( -16.60, z-score = -0.22, R) >droGri2.scaffold_15245 9444183 80 + 18325388 ----ACGCAUUUGGGAUUAGUGCCCAUUAA----------AACACUGAUUGAAGGCCAAAAGUGUG------------CUUUGUCACUUUUCAUUGUUUAUGUUGU------- ----..((((.((((.......))))...(----------((((.(((..((((....((((....------------))))....))))))).)))))))))...------- ( -14.80, z-score = 0.55, R) >consensus ____GCC_CGUUCGGAUUAGGCCUGAUCCA_______CGAGAGCCUGAUUGAAGGUUAACCAUGUGUGAACGUGAGUGUUUUGUUAUGGCUUAUUGUUCGGCGGAU_______ ..............((((((((((...............)).))))))))(((.(.((((((((.....((....)).......))))).))).).))).............. (-11.26 = -12.63 + 1.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:22 2011