
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,143,196 – 3,143,351 |
| Length | 155 |
| Max. P | 0.573615 |

| Location | 3,143,196 – 3,143,351 |
|---|---|
| Length | 155 |
| Sequences | 9 |
| Columns | 173 |
| Reading direction | reverse |
| Mean pairwise identity | 64.08 |
| Shannon entropy | 0.71518 |
| G+C content | 0.38769 |
| Mean single sequence MFE | -35.61 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.60 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.573615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3143196 155 - 21146708 ---UAUAUACGAGGUAUUGGGUGACAAU-AA---AGUUUGUUGCUAGGCCAAAAAGUUCUAGGCAGCCACUU------AGUAAUGGUUUAUGAAAGCAAAAACGUGACAUGAUUGUCACAUGCGUGCGAUUAUCACGUUUUGCGAUAAAGUUUU----CGGAUC-CCAUAACA ---.........(((....((..((((.-..---...))))..))..(((...........))).)))....------.((.((((..(.(((((((.(((((((((..((((((.(((....))))))))))))))))))........)))))----)).)..-)))).)). ( -41.60, z-score = -1.72, R) >droPer1.super_2 1583111 153 - 9036312 -UUCCUCAAUAUAGU---CAAUGAUACACAAUUCGAUUUGUUGCCAGGUCGCAAAAUUUCUGGUCGC----------------AUUAUUAUGAAAGCCGAAACGUGAUAUGAUAGUCACGCGUUUAUGAUUAUCACGUUUUGCGAUAAUUUCCUAAUUUUGGUCUCCAAAACA -..((((((((.(((---((((........))).)))))))))..)))((((((((((((.(((..(----------------((....)))...)))))))((((((((.((((........)))).).)))))))))))))))...........((((((...)))))).. ( -31.70, z-score = -0.54, R) >dp4.chr3 1409889 156 - 19779522 -UUCCUCAAUAUAGUCAGCAAUGAUACACAAUUCGAUUUGUUGCCAGGUCGCAAAAUUUCUGGUCGC----------------AUUAUUAUGAAAGCCGAAACGUGAUAUGAUAGUCACGCGUUUAUGAUUAUCACGUUUUGCGAUAAUUUUCUAAUUUUGAUCUCCAAAACA -............((((....))))........(((((((....))))))).........(((...(----------------(..((((.((((.((((((((((((((.((((........)))).).)))))))))))).)....)))).))))..))....)))..... ( -31.70, z-score = -0.37, R) >droAna3.scaffold_13266 3362635 142 - 19884421 CUUUCCCAAUAACUU---AAAUGACAAACAA----GCUUGUUGCUAGGCCAAAUAAAUUAAGACC-------------------CGAUUAUGAACCAAAAAACGUGAUAUGAUAGUCACAUGUUGGCGAUUAUCACGUUUUGCGAUAACUUCUUA----UGGUC-CCAAAACU ...............---....(((.((.((----(.(((((((..((....((((.........-------------------...))))...))..(((((((((((.....((((.....))))...))))))))))))))))))))).)).----..)))-........ ( -24.90, z-score = -0.78, R) >droEre2.scaffold_4929 7197478 152 + 26641161 ----AUUUAUCCGAUACUGGGGGACACU-AA---AGUUUGUUGCUAGGCCGCAGA-UUCUGGGCAGCCACUU------AGUAAAGGCUUAUGAAAGCGAAAACGUGACAUGAUUGUCGCACGCGUGCGUUUAUCACGUUUGGCGAUAAAGUUU-----CGGAUC-CCAUAACA ----......(((....)))((((..(.-((---(.((((((((((((((.(((.-..))))))((((....------......))))..((((((((...((((((((....))))....)))).))))).)))...))))))))))).)))-----.)..))-))...... ( -44.80, z-score = -0.94, R) >droYak2.chr2L 15835434 141 - 22324452 ----------------------GGCACU-AA---AGUUUGUUGCUAGGCCACAAA-UUUUGGGCAGCCACUUAGUACGAGUAUAGGCUUGUGAAAGCAAAAACGUGACAUGAUUGUCACAUGCGUGCGAUUAUCACGUUUCGCGAUAAAGUUUA----CCGAUC-CCAUAUCA ----------------------(((.((-((---(((((((.((...)).)))))-))))))...)))((((..(((((((....)))))))...((..((((((((..((((((.(((....))))))))))))))))).))....))))...----......-........ ( -40.80, z-score = -1.80, R) >droSec1.super_1 794708 156 - 14215200 ---CAUAUAUGAGAUAUUGGGUGACACUAAA---AGUUUGUUGCCAGGCCAAAAAAUUCUGGGCAGCCACUU------AGUAACGGCUUAUGAAAGCGAAAACGUGACAUGAUUGUCACAUGCGUGCGAUUAUCACGUUUUGCGAUAAAGUUUU----CGGAUC-CCAUAACA ---....((((.(((....(....).(((((---(.((((((((..(.(((........))).)((((....------......))))..........(((((((((..((((((.(((....)))))))))))))))))))))))))).))))----.)))))-.))))... ( -40.60, z-score = -1.01, R) >droSim1.chr2R_random 628971 156 - 2996586 ---CAUAUAUGAGAUAUUGGGUGACACUAAA---AGUUUGUUGCCAGGCCAAAAAAUUCUGGGCAGCCACUU------AGUAACGGCUUAUGAAAGCGAAAACGUGACAUGAUUGUCACAUGCGUGCGAUUAUCACGUUUUGCGAUAAAGUUUU----CGGAUC-CCAUAACA ---....((((.(((....(....).(((((---(.((((((((..(.(((........))).)((((....------......))))..........(((((((((..((((((.(((....)))))))))))))))))))))))))).))))----.)))))-.))))... ( -40.60, z-score = -1.01, R) >droGri2.scaffold_15245 9397229 145 - 18325388 -------UAUAUAGA-UAUAAUGGUAAAAAAAAAAAUUUGUUGCCAGGCCAUAAAAAAUGCACACGUU---------UAGACCUAAAACAUAUGCCCAAAAACGUGAUAUGCCAGUUAGGUACACGCUUUUAUCACGUUU-AUGAUACGCUUAA----UUAAUGACA------ -------........-......(((((.(((.....))).)))))((((..........(((...(((---------(.......))))...))).((.((((((((((((((.....))))........))))))))))-.))....))))..----.........------ ( -23.80, z-score = 0.14, R) >consensus ___CAUAUAUAAGGU_UUGAAUGACACU_AA___AGUUUGUUGCCAGGCCAAAAAAUUUUGGGCAGCCACUU______AGUAAAGGAUUAUGAAAGCAAAAACGUGACAUGAUUGUCACAUGCGUGCGAUUAUCACGUUUUGCGAUAAAGUUUU____CGGAUC_CCAUAACA .....................................(((((((...(((...........)))...................................((((((((..(((((((.........))))))))))))))).)))))))......................... (-14.10 = -14.60 + 0.50)

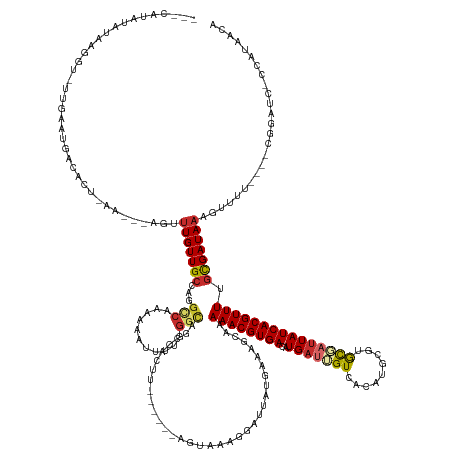
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:16 2011