
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,118,490 – 3,118,615 |
| Length | 125 |
| Max. P | 0.994355 |

| Location | 3,118,490 – 3,118,591 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.49 |
| Shannon entropy | 0.37028 |
| G+C content | 0.41821 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -13.78 |
| Energy contribution | -13.87 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.846281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
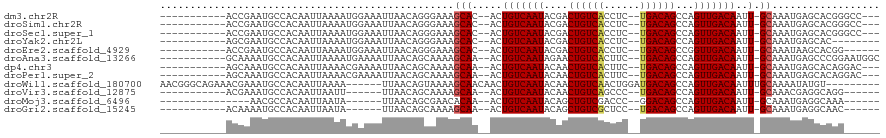
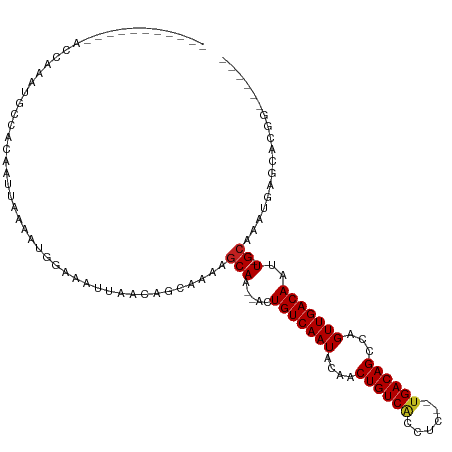
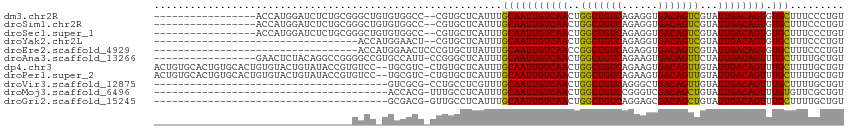
>dm3.chr2R 3118490 101 + 21146708 -----------ACCGAAUGCCACAAUUAAAAUGGAAAUUAACAGGGAAAGCAC--ACUGUCAAUACGACUGUCACCUC--UGACAGCCAGUUGACAAUU-GCAAAUGAGCACGGGCC--- -----------.(((..(((((.......(((....)))..........(((.--..(((((((..(.((((((....--)))))))..)))))))..)-))...)).))))))...--- ( -23.90, z-score = -1.30, R) >droSim1.chr2R 1903333 101 + 19596830 -----------ACCGAAUGCCACAAUUAAAAUGGAAAUUAACAGGGAAAGCAC--ACUGUCAAUACGACUGUCACCUC--UGACAGCCAGUUGACAAUU-GCAAAUGAGCACGGGCC--- -----------.(((..(((((.......(((....)))..........(((.--..(((((((..(.((((((....--)))))))..)))))))..)-))...)).))))))...--- ( -23.90, z-score = -1.30, R) >droSec1.super_1 774900 101 + 14215200 -----------ACCGAAUGCCACAAUUAAAAUGGAAAUUAACAGGGAAAGCAC--ACUGUCAAUACGACUGUCACCUC--UGACAGCCAGUUGACAAUU-GCAAAUGAGCACGGGCC--- -----------.(((..(((((.......(((....)))..........(((.--..(((((((..(.((((((....--)))))))..)))))))..)-))...)).))))))...--- ( -23.90, z-score = -1.30, R) >droYak2.chr2L 15815720 96 + 22324452 -----------AGCGAAUGCCACAAUUAAAAUGGAAAUUAACAGGGAAAGCAC--ACUGUCAAUACGACUGUCACCUC--UGACAGCCAGUUGACAAUU-GCAAAUGAGCAC-------- -----------.((.....(((.........)))...............(((.--..(((((((..(.((((((....--)))))))..)))))))..)-))......))..-------- ( -20.10, z-score = -0.88, R) >droEre2.scaffold_4929 7177408 98 - 26641161 -----------ACCGAAUGCCACAAUUAAAAUGGAAAUUAACAGGGAAAGCAC--ACUGUCAAUACGACUGUCACCUC--UGACAGCCGGUUGACAAUU-GCAAAUAAGCACGG------ -----------.(((..(((.........(((....)))..........(((.--..(((((((.((.((((((....--)))))).)))))))))..)-))......))))))------ ( -24.60, z-score = -2.42, R) >droAna3.scaffold_13266 3343800 104 + 19884421 -----------GCAAAAUGCCACAAUUAAAAUGAAAAUUAACAGCAAAAGCAA--ACUGUCAAUAGAACUGUCACUUC--UGACAGCCAGUUGACAAUU-GCAAAUGAGCCCGGAAUGGC -----------.......((((.......(((....)))..........((((--..(((((((.(..((((((....--)))))).).))))))).))-))..............)))) ( -22.10, z-score = -1.38, R) >dp4.chr3 1388839 101 + 19779522 -----------AGCAAAUGCCACAAUUAAAACGAAAAUUAACAGCAAAAGCAA--ACUGUCAAUACAACUGUCACUUC--UGACAGCCAGUUGACAAUU-GCAAAUGAGCACAGGAC--- -----------.((...(((...((((........))))....)))...((((--..(((((((....((((((....--))))))...))))))).))-))......)).......--- ( -20.90, z-score = -1.76, R) >droPer1.super_2 1563533 101 + 9036312 -----------AGCAAAUGCCACAAUUAAAACGAAAAUUAACAGCAAAAGCAA--ACUGUCAAUACAACUGUCACUUC--UGACAGCCAGUUGACAAUU-GCAAAUGAGCACAGGAC--- -----------.((...(((...((((........))))....)))...((((--..(((((((....((((((....--))))))...))))))).))-))......)).......--- ( -20.90, z-score = -1.76, R) >droWil1.scaffold_180700 2725944 105 + 6630534 AACGGGCAGAAACGAAAUGCCACAAUUAAAA------UUAACAGUAAAAGCAACAACUGUCAAUACAACUGUCAACUGGAUGACAGCCAGUUGACAAUUUGCAAAAUAUGU--------- ....((((.........))))(((......(------((.(((((..........))))).))).....((((((((((.......))))))))))............)))--------- ( -23.90, z-score = -2.48, R) >droVir3.scaffold_12875 18966059 92 - 20611582 -----------ACGAAAUGCCACAAUUAAUU------UUAACAGCAAAAGCAA--ACUGUCAAUACAACUGUCAGCCC--UGACAGCCAGUUGACAAUU-GCAAACGAGGCAGG------ -----------......((((........((------((......))))((((--..(((((((....((((((....--))))))...))))))).))-))......))))..------ ( -23.70, z-score = -2.83, R) >droMoj3.scaffold_6496 22123125 88 - 26866924 ---------------AACGCCACAAUUAAUA------UUAACAGCGAACACAA--ACUGUCAAUACAGCUGUCGACCC--GGACAGCCAGUUGACAAUU-GCAAAUGAGGCAAA------ ---------------...(((..(((....)------))....((((......--..(((((((...((((((.....--.))))))..))))))).))-))......)))...------ ( -21.40, z-score = -2.74, R) >droGri2.scaffold_15245 9371391 92 + 18325388 -----------ACAAAAUGCCACAAUUAAUA------UUAACAGCAAAAGCAA--ACUGUCAAUACAGCUGUCGCUCC--UGACAGCCAGUUGACAAUU-GCAAAUGAGGCAAC------ -----------......((((..(((....)------))..........((((--..(((((((...(((((((....--)))))))..))))))).))-))......))))..------ ( -25.30, z-score = -3.45, R) >consensus ___________ACCAAAUGCCACAAUUAAAAUGGAAAUUAACAGCAAAAGCAA__ACUGUCAAUACAACUGUCACCUC__UGACAGCCAGUUGACAAUU_GCAAAUGAGCACGG______ .................(((.......................(......)......(((((((....((((((......))))))...)))))))....)))................. (-13.78 = -13.87 + 0.10)

| Location | 3,118,490 – 3,118,591 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.49 |
| Shannon entropy | 0.37028 |
| G+C content | 0.41821 |
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -20.77 |
| Energy contribution | -20.48 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
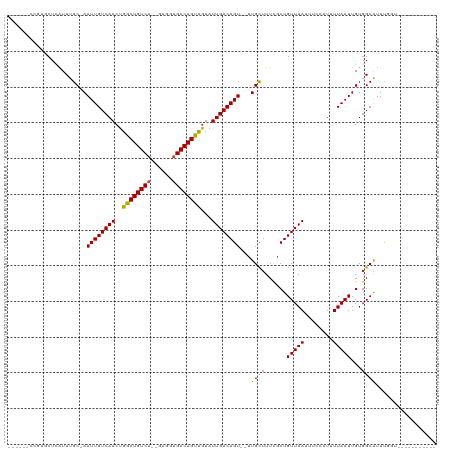
>dm3.chr2R 3118490 101 - 21146708 ---GGCCCGUGCUCAUUUGC-AAUUGUCAACUGGCUGUCA--GAGGUGACAGUCGUAUUGACAGU--GUGCUUUCCCUGUUAAUUUCCAUUUUAAUUGUGGCAUUCGGU----------- ---.(((.((((.(((..((-(((((((((.(((((((((--....)))))))))..))))))))--.))).......(((((........))))).)))))))..)))----------- ( -30.30, z-score = -1.85, R) >droSim1.chr2R 1903333 101 - 19596830 ---GGCCCGUGCUCAUUUGC-AAUUGUCAACUGGCUGUCA--GAGGUGACAGUCGUAUUGACAGU--GUGCUUUCCCUGUUAAUUUCCAUUUUAAUUGUGGCAUUCGGU----------- ---.(((.((((.(((..((-(((((((((.(((((((((--....)))))))))..))))))))--.))).......(((((........))))).)))))))..)))----------- ( -30.30, z-score = -1.85, R) >droSec1.super_1 774900 101 - 14215200 ---GGCCCGUGCUCAUUUGC-AAUUGUCAACUGGCUGUCA--GAGGUGACAGUCGUAUUGACAGU--GUGCUUUCCCUGUUAAUUUCCAUUUUAAUUGUGGCAUUCGGU----------- ---.(((.((((.(((..((-(((((((((.(((((((((--....)))))))))..))))))))--.))).......(((((........))))).)))))))..)))----------- ( -30.30, z-score = -1.85, R) >droYak2.chr2L 15815720 96 - 22324452 --------GUGCUCAUUUGC-AAUUGUCAACUGGCUGUCA--GAGGUGACAGUCGUAUUGACAGU--GUGCUUUCCCUGUUAAUUUCCAUUUUAAUUGUGGCAUUCGCU----------- --------((((.(((..((-(((((((((.(((((((((--....)))))))))..))))))))--.))).......(((((........))))).))))))).....----------- ( -26.90, z-score = -1.78, R) >droEre2.scaffold_4929 7177408 98 + 26641161 ------CCGUGCUUAUUUGC-AAUUGUCAACCGGCUGUCA--GAGGUGACAGUCGUAUUGACAGU--GUGCUUUCCCUGUUAAUUUCCAUUUUAAUUGUGGCAUUCGGU----------- ------((((((......))-)((((((((.(((((((((--....)))))))))..))))))))--(((((......(((((........)))))...))))).))).----------- ( -30.30, z-score = -2.89, R) >droAna3.scaffold_13266 3343800 104 - 19884421 GCCAUUCCGGGCUCAUUUGC-AAUUGUCAACUGGCUGUCA--GAAGUGACAGUUCUAUUGACAGU--UUGCUUUUGCUGUUAAUUUUCAUUUUAAUUGUGGCAUUUUGC----------- (((((....(((......((-(((((((((..((((((((--....)))))).))..))))))).--))))....)))(((((........))))).))))).......----------- ( -30.00, z-score = -2.33, R) >dp4.chr3 1388839 101 - 19779522 ---GUCCUGUGCUCAUUUGC-AAUUGUCAACUGGCUGUCA--GAAGUGACAGUUGUAUUGACAGU--UUGCUUUUGCUGUUAAUUUUCGUUUUAAUUGUGGCAUUUGCU----------- ---((...((((.(((.(((-((((((((.(((.....))--)...)))))))))))((((((((--........))))))))..............)))))))..)).----------- ( -28.60, z-score = -2.12, R) >droPer1.super_2 1563533 101 - 9036312 ---GUCCUGUGCUCAUUUGC-AAUUGUCAACUGGCUGUCA--GAAGUGACAGUUGUAUUGACAGU--UUGCUUUUGCUGUUAAUUUUCGUUUUAAUUGUGGCAUUUGCU----------- ---((...((((.(((.(((-((((((((.(((.....))--)...)))))))))))((((((((--........))))))))..............)))))))..)).----------- ( -28.60, z-score = -2.12, R) >droWil1.scaffold_180700 2725944 105 - 6630534 ---------ACAUAUUUUGCAAAUUGUCAACUGGCUGUCAUCCAGUUGACAGUUGUAUUGACAGUUGUUGCUUUUACUGUUAA------UUUUAAUUGUGGCAUUUCGUUUCUGCCCGUU ---------........(((((.(((((((((((.......))))))))))))))))((((((((..........))))))))------........(.((((.........)))))... ( -29.40, z-score = -3.39, R) >droVir3.scaffold_12875 18966059 92 + 20611582 ------CCUGCCUCGUUUGC-AAUUGUCAACUGGCUGUCA--GGGCUGACAGUUGUAUUGACAGU--UUGCUUUUGCUGUUAA------AAUUAAUUGUGGCAUUUCGU----------- ------..((((.((..(((-((((((((.(((.....))--)...)))))))))))((((((((--........))))))))------.......)).))))......----------- ( -29.40, z-score = -2.74, R) >droMoj3.scaffold_6496 22123125 88 + 26866924 ------UUUGCCUCAUUUGC-AAUUGUCAACUGGCUGUCC--GGGUCGACAGCUGUAUUGACAGU--UUGUGUUCGCUGUUAA------UAUUAAUUGUGGCGUU--------------- ------...(((......((-(((((......(((((((.--.....)))))))(((((((((((--........))))))))------)))))))))))))...--------------- ( -28.10, z-score = -2.77, R) >droGri2.scaffold_15245 9371391 92 - 18325388 ------GUUGCCUCAUUUGC-AAUUGUCAACUGGCUGUCA--GGAGCGACAGCUGUAUUGACAGU--UUGCUUUUGCUGUUAA------UAUUAAUUGUGGCAUUUUGU----------- ------..((((......((-(((((......(((((((.--.....)))))))(((((((((((--........))))))))------))))))))))))))......----------- ( -30.10, z-score = -2.66, R) >consensus ______CCGUGCUCAUUUGC_AAUUGUCAACUGGCUGUCA__GAGGUGACAGUUGUAUUGACAGU__UUGCUUUCGCUGUUAAUUUCCAUUUUAAUUGUGGCAUUUGGU___________ ......................((((((((..((((((((......))))))))...))))))))...((((......(((((........)))))...))))................. (-20.77 = -20.48 + -0.28)


| Location | 3,118,519 – 3,118,615 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 67.44 |
| Shannon entropy | 0.59886 |
| G+C content | 0.49960 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -16.18 |
| Energy contribution | -16.29 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.988338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
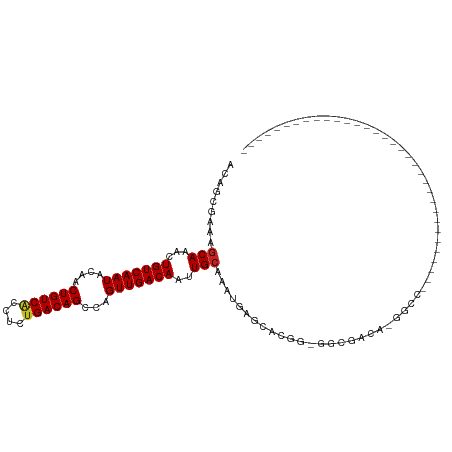
>dm3.chr2R 3118519 96 + 21146708 ACAGGGAAAGCACACUGUCAAUACGACUGUCACCUCUGACAGCCAGUUGACAAUUGCAAAUGAGCACG--GGCCACACAGCCCGCAGAGAUCCAUGGU----------------- .((.(((..(((...(((((((..(.((((((....)))))))..)))))))..))).........((--(((......)))))......))).))..----------------- ( -30.50, z-score = -1.92, R) >droSim1.chr2R 1903362 96 + 19596830 ACAGGGAAAGCACACUGUCAAUACGACUGUCACCUCUGACAGCCAGUUGACAAUUGCAAAUGAGCACG--GGCCACACAGCCCGCAGAGAUCCAUGGU----------------- .((.(((..(((...(((((((..(.((((((....)))))))..)))))))..))).........((--(((......)))))......))).))..----------------- ( -30.50, z-score = -1.92, R) >droSec1.super_1 774929 96 + 14215200 ACAGGGAAAGCACACUGUCAAUACGACUGUCACCUCUGACAGCCAGUUGACAAUUGCAAAUGAGCACG--GGCCACACAGCCCGCAGAGAUCCAUGGU----------------- .((.(((..(((...(((((((..(.((((((....)))))))..)))))))..))).........((--(((......)))))......))).))..----------------- ( -30.50, z-score = -1.92, R) >droYak2.chr2L 15815749 79 + 22324452 ACAGGGAAAGCACACUGUCAAUACGACUGUCACCUCUGACAGCCAGUUGACAAUUGCAAAUGAGCACG--AGUUCCAUGGU---------------------------------- .((.((((..(....(((((((..(.((((((....)))))))..)))))))..(((......))).)--..)))).))..---------------------------------- ( -22.90, z-score = -1.51, R) >droEre2.scaffold_4929 7177437 81 - 26641161 ACAGGGAAAGCACACUGUCAAUACGACUGUCACCUCUGACAGCCGGUUGACAAUUGCAAAUAAGCACGGGAGUUCCAUGGU---------------------------------- .((.((((..(....(((((((.((.((((((....)))))).)))))))))..(((......)))...)..)))).))..---------------------------------- ( -25.50, z-score = -2.08, R) >droAna3.scaffold_13266 3343829 97 + 19884421 ACAGCAAAAGCAAACUGUCAAUAGAACUGUCACUUCUGACAGCCAGUUGACAAUUGCAAAUGAGCCCGG-AAUGGCACGGCCCCGGCCUGUAGAGUUC----------------- ((((.....((((..(((((((.(..((((((....)))))).).))))))).))))........((((-...(((...))))))).)))).......----------------- ( -28.00, z-score = -1.31, R) >dp4.chr3 1388868 112 + 19779522 ACAGCAAAAGCAAACUGUCAAUACAACUGUCACUUCUGACAGCCAGUUGACAAUUGCAAAUGAGCACAG-GACGCA--GGACACGGUAUACAGUACACAGUGCACAGUGCACAGU ...(((...((((..(((((((....((((((....))))))...))))))).))))...((.((((..-..((..--.....))((((...))))...)))).)).)))..... ( -28.20, z-score = -0.31, R) >droPer1.super_2 1563562 112 + 9036312 ACAGCAAAAGCAAACUGUCAAUACAACUGUCACUUCUGACAGCCAGUUGACAAUUGCAAAUGAGCACAG-GACGCA--GGACACGGUAUACAGUACACAGUGCACAGUGCACAGU ...(((...((((..(((((((....((((((....))))))...))))))).))))...((.((((..-..((..--.....))((((...))))...)))).)).)))..... ( -28.20, z-score = -0.31, R) >droVir3.scaffold_12875 18966082 75 - 20611582 ACAGCAAAAGCAAACUGUCAAUACAACUGUCAGCCCUGACAGCCAGUUGACAAUUGCAAACGAGGCAGG-CGCGAC--------------------------------------- ...((....((((..(((((((....((((((....))))))...))))))).))))...(......).-.))...--------------------------------------- ( -20.10, z-score = -1.24, R) >droMoj3.scaffold_6496 22123144 75 - 26866924 ACAGCGAACACAAACUGUCAAUACAGCUGUCGACCCGGACAGCCAGUUGACAAUUGCAAAUGAGGCAAA-CGUGGU--------------------------------------- ........(((....(((((((...((((((......))))))..))))))).((((.......)))).-.)))..--------------------------------------- ( -22.90, z-score = -2.14, R) >droGri2.scaffold_15245 9371414 75 + 18325388 ACAGCAAAAGCAAACUGUCAAUACAGCUGUCGCUCCUGACAGCCAGUUGACAAUUGCAAAUGAGGCAAC-CGUCGC--------------------------------------- ...((....((((..(((((((...(((((((....)))))))..))))))).))))......(((...-.)))))--------------------------------------- ( -23.70, z-score = -2.49, R) >consensus ACAGCGAAAGCAAACUGUCAAUACAACUGUCACCUCUGACAGCCAGUUGACAAUUGCAAAUGAGCACGG_GGCGACA_GGCC_________________________________ .........(((...(((((((....((((((....))))))...)))))))..))).......................................................... (-16.18 = -16.29 + 0.11)


| Location | 3,118,519 – 3,118,615 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 67.44 |
| Shannon entropy | 0.59886 |
| G+C content | 0.49960 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -21.44 |
| Energy contribution | -20.96 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.994355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3118519 96 - 21146708 -----------------ACCAUGGAUCUCUGCGGGCUGUGUGGCC--CGUGCUCAUUUGCAAUUGUCAACUGGCUGUCAGAGGUGACAGUCGUAUUGACAGUGUGCUUUCCCUGU -----------------..((.(((.....(((((((....))))--)))........(((((((((((.(((((((((....)))))))))..)))))))).)))..))).)). ( -38.20, z-score = -2.56, R) >droSim1.chr2R 1903362 96 - 19596830 -----------------ACCAUGGAUCUCUGCGGGCUGUGUGGCC--CGUGCUCAUUUGCAAUUGUCAACUGGCUGUCAGAGGUGACAGUCGUAUUGACAGUGUGCUUUCCCUGU -----------------..((.(((.....(((((((....))))--)))........(((((((((((.(((((((((....)))))))))..)))))))).)))..))).)). ( -38.20, z-score = -2.56, R) >droSec1.super_1 774929 96 - 14215200 -----------------ACCAUGGAUCUCUGCGGGCUGUGUGGCC--CGUGCUCAUUUGCAAUUGUCAACUGGCUGUCAGAGGUGACAGUCGUAUUGACAGUGUGCUUUCCCUGU -----------------..((.(((.....(((((((....))))--)))........(((((((((((.(((((((((....)))))))))..)))))))).)))..))).)). ( -38.20, z-score = -2.56, R) >droYak2.chr2L 15815749 79 - 22324452 ----------------------------------ACCAUGGAACU--CGUGCUCAUUUGCAAUUGUCAACUGGCUGUCAGAGGUGACAGUCGUAUUGACAGUGUGCUUUCCCUGU ----------------------------------..((.((((..--..(((......)))((((((((.(((((((((....)))))))))..)))))))).....)))).)). ( -25.80, z-score = -1.69, R) >droEre2.scaffold_4929 7177437 81 + 26641161 ----------------------------------ACCAUGGAACUCCCGUGCUUAUUUGCAAUUGUCAACCGGCUGUCAGAGGUGACAGUCGUAUUGACAGUGUGCUUUCCCUGU ----------------------------------..(((((.....))))).......(((((((((((.(((((((((....)))))))))..)))))))).)))......... ( -28.90, z-score = -3.12, R) >droAna3.scaffold_13266 3343829 97 - 19884421 -----------------GAACUCUACAGGCCGGGGCCGUGCCAUU-CCGGGCUCAUUUGCAAUUGUCAACUGGCUGUCAGAAGUGACAGUUCUAUUGACAGUUUGCUUUUGCUGU -----------------.......((((...(((.(((.......-.))).)))....(((((((((((..((((((((....)))))).))..))))))).)))).....)))) ( -32.10, z-score = -1.29, R) >dp4.chr3 1388868 112 - 19779522 ACUGUGCACUGUGCACUGUGUACUGUAUACCGUGUCC--UGCGUC-CUGUGCUCAUUUGCAAUUGUCAACUGGCUGUCAGAAGUGACAGUUGUAUUGACAGUUUGCUUUUGCUGU ..((.(((((((((((.(((((...))))).))))..--.)))..-..)))).))...(((((((((((..((((((((....))))))))...))))))).))))......... ( -32.30, z-score = -0.32, R) >droPer1.super_2 1563562 112 - 9036312 ACUGUGCACUGUGCACUGUGUACUGUAUACCGUGUCC--UGCGUC-CUGUGCUCAUUUGCAAUUGUCAACUGGCUGUCAGAAGUGACAGUUGUAUUGACAGUUUGCUUUUGCUGU ..((.(((((((((((.(((((...))))).))))..--.)))..-..)))).))...(((((((((((..((((((((....))))))))...))))))).))))......... ( -32.30, z-score = -0.32, R) >droVir3.scaffold_12875 18966082 75 + 20611582 ---------------------------------------GUCGCG-CCUGCCUCGUUUGCAAUUGUCAACUGGCUGUCAGGGCUGACAGUUGUAUUGACAGUUUGCUUUUGCUGU ---------------------------------------(..((.-...))..)((..(((((((((((..((((((((....))))))))...))))))).))))....))... ( -22.50, z-score = -0.55, R) >droMoj3.scaffold_6496 22123144 75 + 26866924 ---------------------------------------ACCACG-UUUGCCUCAUUUGCAAUUGUCAACUGGCUGUCCGGGUCGACAGCUGUAUUGACAGUUUGUGUUCGCUGU ---------------------------------------......-...((..(((....(((((((((..(((((((......)))))))...))))))))).)))...))... ( -21.20, z-score = -0.76, R) >droGri2.scaffold_15245 9371414 75 - 18325388 ---------------------------------------GCGACG-GUUGCCUCAUUUGCAAUUGUCAACUGGCUGUCAGGAGCGACAGCUGUAUUGACAGUUUGCUUUUGCUGU ---------------------------------------((((((-(((((.......)))))))))..(..((((((((.(((....)))...))))))))..).....))... ( -26.70, z-score = -1.81, R) >consensus _________________________________GGCC_UGGCGCC_CCGUGCUCAUUUGCAAUUGUCAACUGGCUGUCAGAGGUGACAGUUGUAUUGACAGUUUGCUUUCGCUGU ..........................................................(((((((((((..(((((((......)))))))...)))))))).)))......... (-21.44 = -20.96 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:13 2011