| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,060,929 – 3,061,052 |
| Length | 123 |
| Max. P | 0.954281 |

| Location | 3,060,929 – 3,061,052 |
|---|---|
| Length | 123 |
| Sequences | 8 |
| Columns | 131 |
| Reading direction | forward |
| Mean pairwise identity | 74.52 |
| Shannon entropy | 0.50997 |
| G+C content | 0.51880 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -14.99 |
| Energy contribution | -14.24 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.954281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
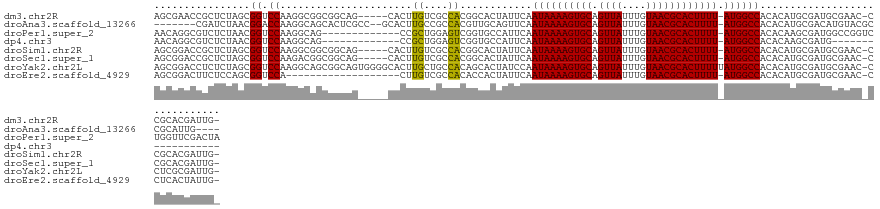
>dm3.chr2R 3060929 123 + 21146708 AGCGAACCGCUCUAGCGGUCCAAGGCGGCGGCAG-----CACUUGUCGCCACGGCACUAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUUUU-AUGGCCACACAUGCGAUGCGAAC-CCGCACGAUUG- .(((.(((((....))))).)..(((((((((((-----...)))))))).............((((((((((.((((....))))))))))))-)).)))......))(.((((...-.))))).....- ( -45.60, z-score = -2.73, R) >droAna3.scaffold_13266 3298611 117 + 19884421 -------CGAUCUAACGGACCAAGGCAGCACUCGCC--GCACUUGCCGCCACGUUGCAGUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUUUU-AUGGCCACACAUGCGACAUGUACGCCGCAUUG---- -------.........((..((((((.((....)).--)).))))...))....(((......((((((((((.((((....))))))))))))-))(((...(((((...)))))..))))))...---- ( -34.80, z-score = -1.19, R) >droPer1.super_2 1525829 117 + 9036312 AACAGGCGUCUCUAACGGUCCAAGGCAG-------------CCGCUGGAGUCGGUGCCAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUUUU-AUGGCCACACAAGCGAUGGCCGGUCUGGUUCGACUA ..(((.((.(((((.((((........)-------------))).))))).))..(((.....((((((((((.((((....))))))))))))-))((((((......).)))))))))))......... ( -41.40, z-score = -2.17, R) >dp4.chr3 1351553 99 + 19779522 AACAGGCGUCUCUAACGGUCCAAGGCAG-------------CCGCUGGAGUCGGUGCCAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUUUU-AUGGCCACACAAGCGAUG------------------ .....(((.(((((.((((........)-------------))).))))).).((((((.....(((((((((.((((....))))))))))))-)))).)))....))....------------------ ( -32.20, z-score = -2.14, R) >droSim1.chr2R 1858637 123 + 19596830 AGCGGACCGCUCUAGCGGUCCAAGGCGGCGGCAG-----CACUUGUCGCCACGGCACUAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUUUU-AUGGCCACACAUGCGAUGCGAAC-CCGCACGAUUG- .(((((((((....)))))))..(((((((((((-----...)))))))).............((((((((((.((((....))))))))))))-)).)))......))(.((((...-.))))).....- ( -52.40, z-score = -4.43, R) >droSec1.super_1 728139 123 + 14215200 AGCGGACCGCUCUAGCGGUCCAAGACGGCGGCAG-----CACUUGUCGCCACGGCACUAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUUUU-AUGGCCACACAUGCGAUGCGAAC-CCGCACGAUUG- .(((((((((....))))))).....((((((((-----...))))))))..(((........((((((((((.((((....))))))))))))-)).)))......))(.((((...-.))))).....- ( -50.60, z-score = -4.49, R) >droYak2.chr2L 15769703 129 + 22324452 AGCGGACCUCUCUAGCGGUCCAAGGCAGCGGCAGUGGGGCACUUGCUGCCACAGCACUAUCCAAUAAAAGUGCAGUUAUUUGUAACGCACUUUUUAUGGCCACACAUGCGAUGCGAAC-CCUCGCGAUUG- .(((((((.(....).)))))..(((.(.(((((..((...))..))))).).........((..((((((((.((((....))))))))))))..)))))......))..(((((..-..)))))....- ( -47.30, z-score = -2.31, R) >droEre2.scaffold_4929 7132592 109 - 26641161 AGCGGACUUCUCCAGCGGUCCA-------------------CUUGUCGCCACACCACUAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUUUU-AUGGCCACACAUGCGAUGCGAAC-CCUCACUAUUG- .(((((((.(....).))))).-------------------...(((((..............((((((((((.((((....))))))))))))-))(......)..)))))))....-...........- ( -27.70, z-score = -2.31, R) >consensus AGCGGACCGCUCUAGCGGUCCAAGGCAGCGGCAG_____CACUUGUCGCCACGGCACUAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUUUU_AUGGCCACACAUGCGAUGCGAAC_CCGCACGAUUG_ ................((.(((.....................((....))..............((((((((.((((....))))))))))))..))))).............................. (-14.99 = -14.24 + -0.75)
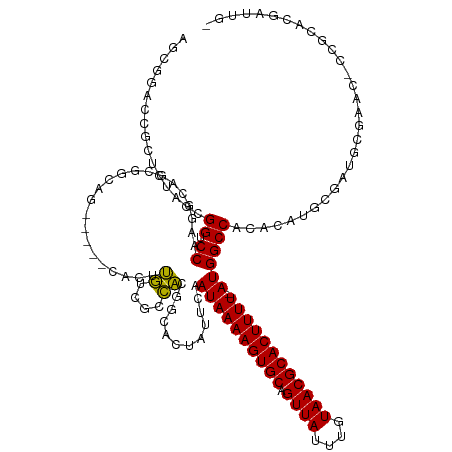
| Location | 3,060,929 – 3,061,052 |
|---|---|
| Length | 123 |
| Sequences | 8 |
| Columns | 131 |
| Reading direction | reverse |
| Mean pairwise identity | 74.52 |
| Shannon entropy | 0.50997 |
| G+C content | 0.51880 |
| Mean single sequence MFE | -42.73 |
| Consensus MFE | -16.67 |
| Energy contribution | -16.31 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.645889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3060929 123 - 21146708 -CAAUCGUGCGG-GUUCGCAUCGCAUGUGUGGCCAU-AAAAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUAGUGCCGUGGCGACAAGUG-----CUGCCGCCGCCUUGGACCGCUAGAGCGGUUCGCU -....(((((((-.......))))))).(((((.((-((((((((((((....)))).))))))))))....(((..(..((....)).)..-----)))..)))))...(((((((....)))))))... ( -46.30, z-score = -1.42, R) >droAna3.scaffold_13266 3298611 117 - 19884421 ----CAAUGCGGCGUACAUGUCGCAUGUGUGGCCAU-AAAAGUGCGUUACAAAUAACUGCACUUUUAUUGAACUGCAACGUGGCGGCAAGUGC--GGCGAGUGCUGCCUUGGUCCGUUAGAUCG------- ----......(((((((.((((((((.(((.(((((-((((((((((((....)))).)))))))).(((.....))).))))).))).))))--)))).)))).)))..((((.....)))).------- ( -47.90, z-score = -2.82, R) >droPer1.super_2 1525829 117 - 9036312 UAGUCGAACCAGACCGGCCAUCGCUUGUGUGGCCAU-AAAAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUGGCACCGACUCCAGCGG-------------CUGCCUUGGACCGUUAGAGACGCCUGUU ..(((......))).(((((.((....)))))))((-((((((((((((....)))).)))))))))).....(((.....(((.(((((-------------((.....)).))))).)))..))).... ( -37.70, z-score = -1.99, R) >dp4.chr3 1351553 99 - 19779522 ------------------CAUCGCUUGUGUGGCCAU-AAAAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUGGCACCGACUCCAGCGG-------------CUGCCUUGGACCGUUAGAGACGCCUGUU ------------------....((..((((.(((((-((((((((((((....)))).))))))))).....)))).....(((.(((((-------------((.....)).))))).)))))))..)). ( -31.40, z-score = -1.95, R) >droSim1.chr2R 1858637 123 - 19596830 -CAAUCGUGCGG-GUUCGCAUCGCAUGUGUGGCCAU-AAAAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUAGUGCCGUGGCGACAAGUG-----CUGCCGCCGCCUUGGACCGCUAGAGCGGUCCGCU -....(((((((-.......))))))).(((((.((-((((((((((((....)))).))))))))))....(((..(..((....)).)..-----)))..)))))...(((((((....)))))))... ( -49.20, z-score = -2.21, R) >droSec1.super_1 728139 123 - 14215200 -CAAUCGUGCGG-GUUCGCAUCGCAUGUGUGGCCAU-AAAAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUAGUGCCGUGGCGACAAGUG-----CUGCCGCCGUCUUGGACCGCUAGAGCGGUCCGCU -.....(((((.-...))))).(((((.(((((.((-((((((((((((....)))).)))))))))).....((..(..((....)).)..-----))))))))))...(((((((....))))))))). ( -48.60, z-score = -2.28, R) >droYak2.chr2L 15769703 129 - 22324452 -CAAUCGCGAGG-GUUCGCAUCGCAUGUGUGGCCAUAAAAAGUGCGUUACAAAUAACUGCACUUUUAUUGGAUAGUGCUGUGGCAGCAAGUGCCCCACUGCCGCUGCCUUGGACCGCUAGAGAGGUCCGCU -.....(((...-...)))...(((.(..(..(((..((((((((((((....)))).))))))))..)))...)..))))((((((.((((...))))...))))))..(((((........)))))... ( -46.70, z-score = -1.18, R) >droEre2.scaffold_4929 7132592 109 + 26641161 -CAAUAGUGAGG-GUUCGCAUCGCAUGUGUGGCCAU-AAAAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUAGUGGUGUGGCGACAAG-------------------UGGACCGCUGGAGAAGUCCGCU -...........-((.((((((((.(((....(.((-((((((((((((....)))).)))))))))).).)))))))))))))....((-------------------(((((..(....)..))))))) ( -34.00, z-score = -1.36, R) >consensus _CAAUCGUGCGG_GUUCGCAUCGCAUGUGUGGCCAU_AAAAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUAGUGCCGUGGCGACAAGUG_____CUGCCGCUGCCUUGGACCGCUAGAGAGGUCCGCU ............................(((((((..((((((((((((....)))).))))))))..............((....)).....................))).)))).............. (-16.67 = -16.31 + -0.36)

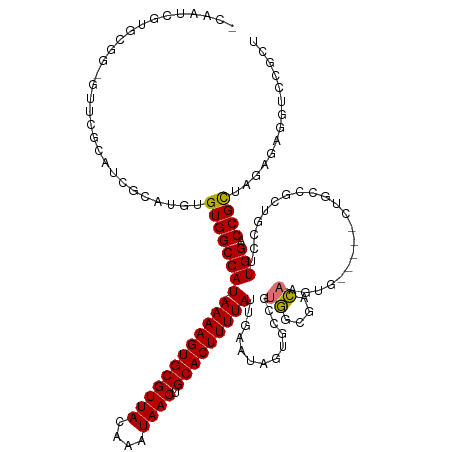
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:07 2011