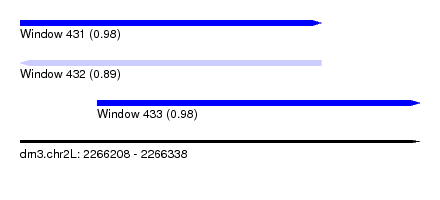
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,266,208 – 2,266,338 |
| Length | 130 |
| Max. P | 0.984834 |

| Location | 2,266,208 – 2,266,306 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.68 |
| Shannon entropy | 0.39411 |
| G+C content | 0.44105 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -18.81 |
| Energy contribution | -19.67 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
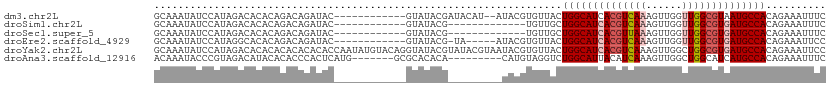
>dm3.chr2L 2266208 98 + 23011544 GCAAAUAUCCAUAGACACACAGACAGAUAC------------GUAUACGAUACAU--AUACGUGUUACUGGCAUCACGUCAAAGUUGGUUGGCGUAAUGCCACAGAAAUUUC .........................(((((------------(((((.......)--)))))))))..((((((.(((((((......))))))).)))))).......... ( -25.80, z-score = -2.68, R) >droSim1.chr2L 2224939 87 + 22036055 GCAAAUAUCCAUAGACACACAGACAGAUAC------------GUAUACG-------------UGUUGCUGGCAUCACGUCAAAGUUGGUUGGCGUGAUGCCACAGAAAUUUC ....................((...(((((------------(....))-------------)))).))(((((((((((((......)))))))))))))........... ( -28.60, z-score = -3.18, R) >droSec1.super_5 441109 87 + 5866729 GCAAAUAUCCAUAGACACACAGACAGAUAC------------GUAUACG-------------UGUUGCUGGCAUCACGUUAAAGUUGGUUGGCGUGAUGCCACAGAAAUUUC ....................((...(((((------------(....))-------------)))).))(((((((((((((......)))))))))))))........... ( -26.50, z-score = -2.65, R) >droEre2.scaffold_4929 2303370 94 + 26641161 GCAAAUAUCCAUAGGCACACAGACAGAUAC------------GUAUACG-UA-----AUACGUGUUACUGGCAUCACGUCAAAGUUGGUUGGCGUGAUGCCACAGAAAUUCC .........................(((((------------((((...-..-----)))))))))..((((((((((((((......)))))))))))))).......... ( -31.40, z-score = -3.65, R) >droYak2.chr2L 2247316 112 + 22324452 GCAAAUAUCCAUAGACACACACACACACACCAAUAUGUACAGGUAUACGUAUACGUAAUACGUGUUACUGGCAUCACGUCAAAGUUGGCUGGCGUGAUGCCACAGAAAUUCC .............(((((........(((......)))....(((((((....))).)))))))))..(((((((((((((........))))))))))))).......... ( -32.10, z-score = -2.45, R) >droAna3.scaffold_12916 9981664 96 - 16180835 ACAAAUACCCGUAGACAUACACACCCACUCAUG-------GCGCACACA---------CAUGUAGGUCUGGCAUUACAUCAAAGUUGGCUGGCAUCAUGCCACAGAAAUUUC ..........(((....)))....((...((((-------.........---------))))..))((((((...((......))..))((((.....))))))))...... ( -16.00, z-score = 1.07, R) >consensus GCAAAUAUCCAUAGACACACAGACAGAUAC____________GUAUACG_________UACGUGUUACUGGCAUCACGUCAAAGUUGGUUGGCGUGAUGCCACAGAAAUUUC ....................................................................((((((((((((((......)))))))))))))).......... (-18.81 = -19.67 + 0.86)
| Location | 2,266,208 – 2,266,306 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.68 |
| Shannon entropy | 0.39411 |
| G+C content | 0.44105 |
| Mean single sequence MFE | -28.11 |
| Consensus MFE | -20.02 |
| Energy contribution | -19.63 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2266208 98 - 23011544 GAAAUUUCUGUGGCAUUACGCCAACCAACUUUGACGUGAUGCCAGUAACACGUAU--AUGUAUCGUAUAC------------GUAUCUGUCUGUGUGUCUAUGGAUAUUUGC ..........((((((((((.(((......))).))))))))))((((.((((((--(((...)))))))------------))...((((((((....)))))))).)))) ( -29.00, z-score = -2.47, R) >droSim1.chr2L 2224939 87 - 22036055 GAAAUUUCUGUGGCAUCACGCCAACCAACUUUGACGUGAUGCCAGCAACA-------------CGUAUAC------------GUAUCUGUCUGUGUGUCUAUGGAUAUUUGC ..........((((((((((.(((......))).))))))))))((((.(-------------((....)------------))...((((((((....)))))))).)))) ( -26.50, z-score = -2.32, R) >droSec1.super_5 441109 87 - 5866729 GAAAUUUCUGUGGCAUCACGCCAACCAACUUUAACGUGAUGCCAGCAACA-------------CGUAUAC------------GUAUCUGUCUGUGUGUCUAUGGAUAUUUGC ..........((((((((((..............))))))))))((((.(-------------((....)------------))...((((((((....)))))))).)))) ( -23.34, z-score = -1.65, R) >droEre2.scaffold_4929 2303370 94 - 26641161 GGAAUUUCUGUGGCAUCACGCCAACCAACUUUGACGUGAUGCCAGUAACACGUAU-----UA-CGUAUAC------------GUAUCUGUCUGUGUGCCUAUGGAUAUUUGC .((((.((((((((((((((.(((......))).)))))))))((..(((((.((-----((-((....)------------)))...)).)))))..))))))).)))).. ( -29.20, z-score = -2.36, R) >droYak2.chr2L 2247316 112 - 22324452 GGAAUUUCUGUGGCAUCACGCCAGCCAACUUUGACGUGAUGCCAGUAACACGUAUUACGUAUACGUAUACCUGUACAUAUUGGUGUGUGUGUGUGUGUCUAUGGAUAUUUGC .((((.((..((((((((((.(((......))).))))))))))......((((.((((((((((((((((..........))))))))))))))))..)))))).)))).. ( -36.20, z-score = -1.42, R) >droAna3.scaffold_12916 9981664 96 + 16180835 GAAAUUUCUGUGGCAUGAUGCCAGCCAACUUUGAUGUAAUGCCAGACCUACAUG---------UGUGUGCGC-------CAUGAGUGGGUGUGUAUGUCUACGGGUAUUUGU .((((.(((((((((((......((..((......))...))((.((((((.((---------((.(....)-------)))).)))))).)))))).))))))).)))).. ( -24.40, z-score = 0.75, R) >consensus GAAAUUUCUGUGGCAUCACGCCAACCAACUUUGACGUGAUGCCAGUAACACGUA_________CGUAUAC____________GUAUCUGUCUGUGUGUCUAUGGAUAUUUGC ..........((((((((((.(((......))).))))))))))......................................(((..((((((((....))))))))..))) (-20.02 = -19.63 + -0.38)
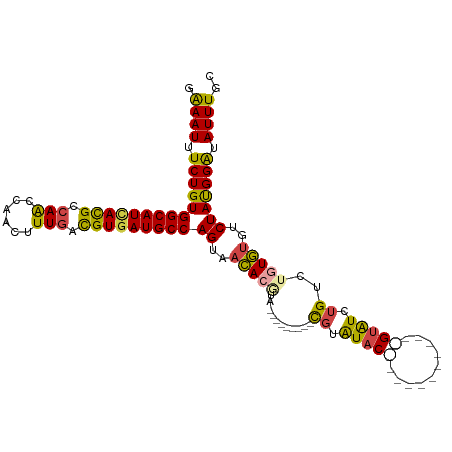
| Location | 2,266,233 – 2,266,338 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.85 |
| Shannon entropy | 0.62446 |
| G+C content | 0.46344 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -11.37 |
| Energy contribution | -11.69 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.978938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2266233 105 + 23011544 GAUACGUAUACG-AUACAUA-UACGUGUUACUGGCAUCACGUCAAAGUUG-GUUGGCGUAAUGCCACAGAAAUUUCAGCAGCCUGCAACAA--------AAGCGAGUCAACCAAAA ((((((((((..-.....))-))))))))..((((((.(((((((.....-.))))))).))))))..............((.(((.....--------..))).))......... ( -27.20, z-score = -1.73, R) >droSim1.chr2L 2224964 94 + 22036055 -----------G-AUACGUA-UACGUGUUGCUGGCAUCACGUCAAAGUUG-GUUGGCGUGAUGCCACAGAAAUUUCAGCUGCCUGCAACAA--------AAGCGAGUCAGCCAAAA -----------(-((.(((.-....((((((((((((((((((((.....-.))))))))))))))(((.........)))...)))))).--------..))).)))........ ( -36.10, z-score = -3.51, R) >droSec1.super_5 441134 94 + 5866729 -----------G-AUACGUA-UACGUGUUGCUGGCAUCACGUUAAAGUUG-GUUGGCGUGAUGCCACAGAAAUUUCAGCUGCCUGCAACAA--------AAGCGAGUCAGCCAGAA -----------(-((.(((.-....((((((((((((((((((((.....-.))))))))))))))(((.........)))...)))))).--------..))).)))........ ( -34.00, z-score = -2.76, R) >droEre2.scaffold_4929 2303395 101 + 26641161 ------GAUACGUAUACGUAAUACGUGUUACUGGCAUCACGUCAAAGUUG-GUUGGCGUGAUGCCACAGAAAUUCCAGCAGGCUGCAACAA--------AAGCGAGUCAACUGAAA ------(((((((((.....)))))))))..((((((((((((((.....-.)))))))))))))).........(((..((((((.....--------..)).))))..)))... ( -39.00, z-score = -4.59, R) >droYak2.chr2L 2247356 104 + 22324452 ---AGGUAUACGUAUACGUAAUACGUGUUACUGGCAUCACGUCAAAGUUG-GCUGGCGUGAUGCCACAGAAAUUCCCGCUGGCUGCAGCAA--------AAGCAAGUCAACUGAAA ---.(((((((((((.....))))))).))))((((((((((((......-..)))))))))))).(((........(((.((....))..--------.))).......)))... ( -36.66, z-score = -2.76, R) >droAna3.scaffold_12916 9981689 97 - 16180835 ---CACUCAUGGCGCACACA-CAUGUAGGUCUGGCAUUACAUCAAAGUUG-GCUGGCAUCAUGCCACAGAAAUUUC------CUGAAACAA--------AAGCAAGUCGACAAAAA ---..((((((.........-)))).))..................((((-((((((.....))).(((.......------)))......--------.....)))))))..... ( -18.60, z-score = 0.51, R) >droPer1.super_1 4638714 101 - 10282868 --------------CACACA-CACACUCGCUGUGUGGCACGUCAAAGUUGGCCUGGCAUCGUGCCACUGGAAUUUCCGUUGGCCACAUCACCUUCCUUCACACACAACAGCAAAAA --------------......-.......((((((((((..((((....))))...))...((((((.(((.....))).))).)))................))).)))))..... ( -27.10, z-score = -1.00, R) >droWil1.scaffold_180708 2215471 82 + 12563649 ------------CACAGGCG-UAAAU-UUUCUGGCGUCAUGUCAAAGUUGUGUUGGCAGCAUGCCACAAAAAUUUA------CUCCAACAA--------G-GAGAAACAAC----- ------------....((.(-(((((-(((.((((((..((((((.......))))))..))))))..))))))))------).)).....--------.-..........----- ( -23.20, z-score = -2.46, R) >consensus ___________G_AUACGUA_UACGUGUUACUGGCAUCACGUCAAAGUUG_GUUGGCGUGAUGCCACAGAAAUUUCAGCUGCCUGCAACAA________AAGCGAGUCAACCAAAA ...............................((((((.((((((.........)))))).)))))).................................................. (-11.37 = -11.69 + 0.31)
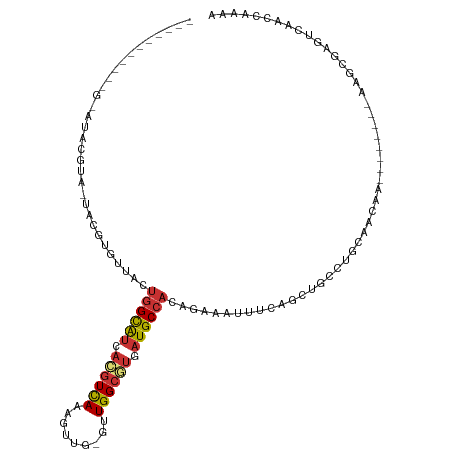
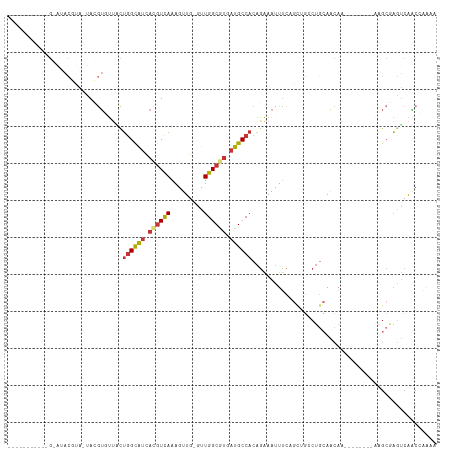
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:40 2011