| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,033,408 – 3,033,498 |
| Length | 90 |
| Max. P | 0.983530 |

| Location | 3,033,408 – 3,033,498 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.15 |
| Shannon entropy | 0.36213 |
| G+C content | 0.47841 |
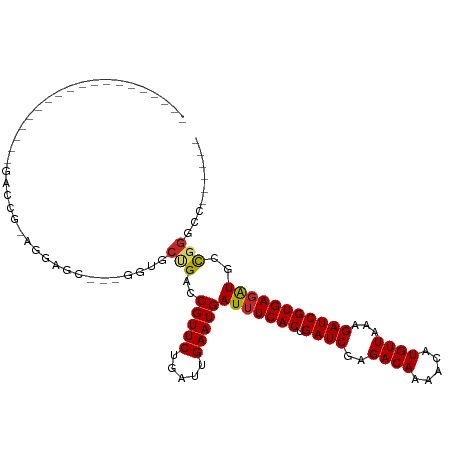
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -16.99 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 3033408 90 - 21146708 -------------------GACCG-AGGAGU---GGUGCUGACCGUUCUGAUUGAAUGAUUUCACUGAUCGAGACAAAACAUGUUAAAGAUCGUGAGAUGCCGGUGAGAUGGA -------------------.((((-.(((((---(((....)))))))).........(((((((.((((..((((.....))))...)))))))))))..))))........ ( -24.80, z-score = -1.59, R) >droAna3.scaffold_13266 3272617 85 - 19884421 -------------------GACCGAAGAAGC---GACGCUGACCGUUCUGAUUGAAUGAUUUCACUGAUCGAGACAAAACAUGUUAAAGAUCGUGAGAUGCCCGACC------ -------------------..........((---.........(((((.....)))))(((((((.((((..((((.....))))...)))))))))))))......------ ( -18.60, z-score = -1.73, R) >droEre2.scaffold_4929 7105481 84 + 26641161 -------------------GACCG-AGGAGC---GGUGCUGACCGUUCUGAUUGAAUGAUUUCACUGAUCGAGACAAAACAUGUUAAAGAUCGUGAGAUGCCGGGCC------ -------------------..(((-.(((((---(((....)))))))).........(((((((.((((..((((.....))))...)))))))))))..)))...------ ( -28.60, z-score = -3.14, R) >droYak2.chr2L 15742276 104 - 22324452 GACAGUGGACAGAGGAUAGAGGAGCGGAGCU---GGUGCUGACCGUUCUGAUUGAAUGAUUUCACUGAUCGAGACAAAACAUGUUAAAGAUCGUGAGAUGCCGGGCC------ .....(((.((...(((...((((((((((.---...)))..))))))).)))...))(((((((.((((..((((.....))))...))))))))))).)))....------ ( -28.90, z-score = -1.22, R) >droSec1.super_1 701246 90 - 14215200 -------------------GACCG-AGGAGC---GGUGCUGACCGUUCUGAUUGAAUGAUUUCACUGAUCGAGACAAAACAUGUUAAAGAUCGUGAGAUGCCGGUGAGAUGCU -------------------.((((-.(((((---(((....)))))))).........(((((((.((((..((((.....))))...)))))))))))..))))........ ( -29.60, z-score = -3.16, R) >droSim1.chr2R 1831508 90 - 19596830 -------------------GACCG-AGGAGC---GGUGCUGACCGUUCUGAUUGAAUGAUUUCACUGAUCGAGACAAAACAUGUUAAAGAUCGUGAGAUGCCGGUGAGAUGCU -------------------.((((-.(((((---(((....)))))))).........(((((((.((((..((((.....))))...)))))))))))..))))........ ( -29.60, z-score = -3.16, R) >droVir3.scaffold_10324 1204165 87 - 1288806 -------------------GACCGGAGCUGCCAAAGCGCUGACCGUUCUGAUUGAAUGAUUUCACUGAUCGAGACAAAACAUGUUAAAGAUCGUGAUGUUGUUGCU------- -------------------((.((((((.((....)))))..))).))......((..((.((((.((((..((((.....))))...)))))))).))..))...------- ( -21.10, z-score = -1.04, R) >droPer1.super_2 1494243 84 - 9036312 -------------------GACCG-CAGCCC---GAUGCUGACCGUUCUGAUUGAAUGAUUUCACUGAUCGAGACAAAACAUGUUAAAGAUCGUGAGAUGCCGGGCC------ -------------------.....-..((((---(..((....(((((.....)))))(((((((.((((..((((.....))))...)))))))))))))))))).------ ( -27.70, z-score = -3.61, R) >dp4.chr3 1319888 84 - 19779522 -------------------GACCG-CAGCCC---GAUGCUGACCGUUCUGAUUGAAUGAUUUCACUGAUCGAGACAAAACAUGUUAAAGAUCGUGAGAUGCCGGGCC------ -------------------.....-..((((---(..((....(((((.....)))))(((((((.((((..((((.....))))...)))))))))))))))))).------ ( -27.70, z-score = -3.61, R) >droWil1.scaffold_180700 2638040 79 - 6630534 -------------------------GGUCCU---GGCAACGACCGUUCUGAUUGAAUGAUUUCACUGAUCGAGACAAAACAUGUUAAAGAUCGUGAGAUGUUGGACC------ -------------------------(((((.---(....)((((((((.....)))))(((((((.((((..((((.....))))...)))))))))))))))))))------ ( -26.10, z-score = -3.16, R) >consensus ___________________GACCG_AGGAGC___GGUGCUGACCGUUCUGAUUGAAUGAUUUCACUGAUCGAGACAAAACAUGUUAAAGAUCGUGAGAUGCCGGGCC______ ......................................(((..(((((.....)))))(((((((.((((..((((.....))))...)))))))))))..)))......... (-16.99 = -17.07 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:04 2011