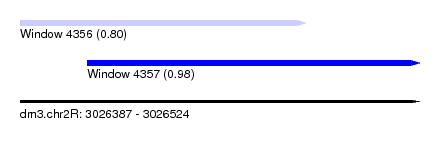
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,026,387 – 3,026,524 |
| Length | 137 |
| Max. P | 0.975371 |

| Location | 3,026,387 – 3,026,485 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.31 |
| Shannon entropy | 0.30192 |
| G+C content | 0.43127 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.18 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797071 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
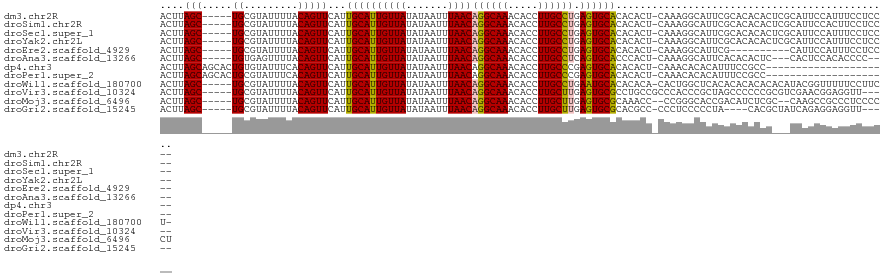
>dm3.chr2R 3026387 98 + 21146708 -----------GAGCGGGAGCUCUAAGGAGCAGCACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACA -----------((((....))))......(((((.....))-----)))................((((((((((.......))))((((((.....)))))).)))))).... ( -30.30, z-score = -2.26, R) >droSim1.chr2R 1825551 98 + 19596830 -----------GAGCGGGAGCUCUAAGGAGCAGCACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACA -----------((((....))))......(((((.....))-----)))................((((((((((.......))))((((((.....)))))).)))))).... ( -30.30, z-score = -2.26, R) >droSec1.super_1 695607 98 + 14215200 -----------GAGCGGGAGCUCUAAGGAGCAGCACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACA -----------((((....))))......(((((.....))-----)))................((((((((((.......))))((((((.....)))))).)))))).... ( -30.30, z-score = -2.26, R) >droYak2.chr2L 15736480 98 + 22324452 -----------CGGCGGGAGCUCUAAGGAGCAGCACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACA -----------......(((((.(((((.(((((.....))-----)))...))))).)))))..((((((((((.......))))((((((.....)))))).)))))).... ( -29.80, z-score = -1.98, R) >droEre2.scaffold_4929 7099627 98 - 26641161 -----------UGGCUGGAGCUCUAAGGAGCAGCACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACA -----------......(((((.(((((.(((((.....))-----)))...))))).)))))..((((((((((.......))))((((((.....)))))).)))))).... ( -29.80, z-score = -1.67, R) >droAna3.scaffold_13266 3267520 108 + 19884421 GAGCGGGCACUAAGC-CAAGUUCUGAGGAGCAGCACUUAGC-----UGUGAGUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUCAGUGCACCCA ..(.(((((((.((.-((((..(((..((((..(((.....-----.))).))))..)))....((((.((((((.......))))))))))....)))).)).))))).))). ( -30.80, z-score = -1.17, R) >dp4.chr3 1314797 104 + 19779522 ----------UUAAGGAGAGCACUAAGGAGCAGCACUUAGCAGCACUGUGUAUUUCACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCCGAGUGCACACA ----------.........(((((..((.(((((.....)).(.((((((.....)))))).).((((.((((((.......))))))))))......))))).)))))..... ( -29.20, z-score = -2.35, R) >droPer1.super_2 1489180 104 + 9036312 ----------UUAAGGAGAGCACUAAGGAGCAGCACUUAGCAGCACUGCGUAUUUCACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCCGAGUGCACACA ----------.........(((((..((.(((((.....)).(.((((.(.....).)))).).((((.((((((.......))))))))))......))))).)))))..... ( -25.30, z-score = -1.22, R) >droWil1.scaffold_180700 2634227 104 + 6630534 -----UGCAUUAAGCACUAAGCAGAAGUAGCAGCACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAAUGCACACA -----((((((..(((.(.(((.(((((((((((.....))-----))).))))))...))).).))).................(((((((.....))))))))))))).... ( -30.40, z-score = -2.67, R) >droVir3.scaffold_10324 1198695 94 + 1288806 ---------------CUGCACACUGAGCAGCAGCACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCUUGAGUGCGCCUG ---------------..((.((((((((((((((.....))-----)))...............((((.((((((.......))))))))))......))))).)))).))... ( -29.40, z-score = -2.10, R) >droMoj3.scaffold_6496 22021896 94 - 26866924 ---------------CUGCGCACUAAGCAGCAGCACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCUUGAGUGCGCAAA ---------------.((((((((((((((((((.....))-----)))...............((((.((((((.......))))))))))......))))).)))))))).. ( -36.40, z-score = -4.51, R) >droGri2.scaffold_15245 9281252 94 + 18325388 ---------------CUGCGCACUAAGCAGCAGCACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCUUGAGUGCGCACG ---------------.((((((((((((((((((.....))-----)))...............((((.((((((.......))))))))))......))))).)))))))).. ( -36.10, z-score = -4.51, R) >consensus ____________AGCGGGAGCUCUAAGGAGCAGCACUUAGC_____UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACA ................................(((((............((((...(((((.......))))).)))).......(((((((.....))))))))))))..... (-17.02 = -17.18 + 0.15)
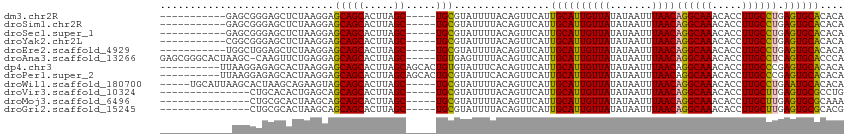
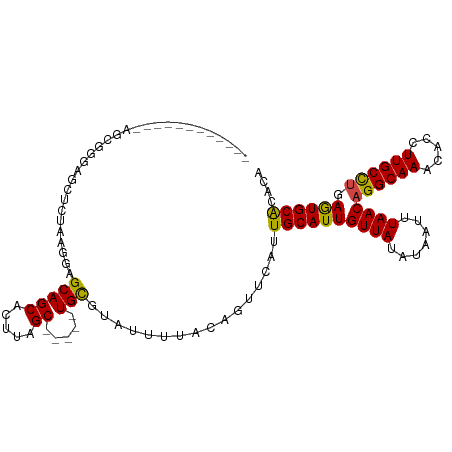
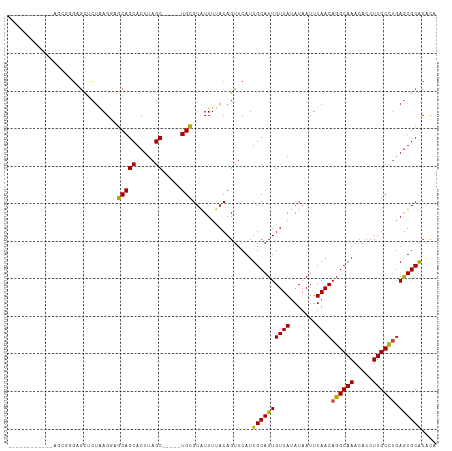
| Location | 3,026,410 – 3,026,524 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 76.09 |
| Shannon entropy | 0.52184 |
| G+C content | 0.43747 |
| Mean single sequence MFE | -23.51 |
| Consensus MFE | -13.16 |
| Energy contribution | -12.88 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
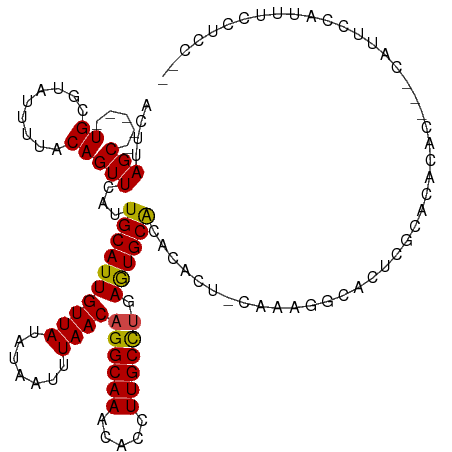
>dm3.chr2R 3026410 114 + 21146708 ACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACACU-CAAAGGCAUUCGCACACACUCGCAUUCCAUUUCCUCC-- .....(.-----((((..............((((.((((((.......))))))))))......((((((((((....))))-)..)))))..)))))......................-- ( -24.10, z-score = -2.39, R) >droSim1.chr2R 1825574 114 + 19596830 ACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACACU-CAAAGGCAUUCGCACACACUCGCAUUCCACUUCCUCC-- .....(.-----((((..............((((.((((((.......))))))))))......((((((((((....))))-)..)))))..)))))......................-- ( -24.10, z-score = -2.34, R) >droSec1.super_1 695630 114 + 14215200 ACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACACU-CAAAGGCAUUCGCACACACUCGCAUUCCAUUUCCUCC-- .....(.-----((((..............((((.((((((.......))))))))))......((((((((((....))))-)..)))))..)))))......................-- ( -24.10, z-score = -2.39, R) >droYak2.chr2L 15736503 114 + 22324452 ACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACACU-CAAAGGCAUUCGCACACACUCGCAUUCCAUUUCCUCC-- .....(.-----((((..............((((.((((((.......))))))))))......((((((((((....))))-)..)))))..)))))......................-- ( -24.10, z-score = -2.39, R) >droEre2.scaffold_4929 7099650 104 - 26641161 ACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACACU-CAAAGGCAUUCG----------CAUUCCAUUUCCUCC-- .......-----((((..............((((.((((((.......))))))))))......((((((((((....))))-)..)))))..))----------)).............-- ( -24.00, z-score = -2.85, R) >droAna3.scaffold_13266 3267553 109 + 19884421 ACUUAGC-----UGUGAGUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUCAGUGCACCCACU-CAAAGGCAUUCACACACUC---CACUCCACACCCC---- .....(.-----(((((((...........((((.((((((.......)))))))))).......((((.((((....))))-...))))))))))))....---.............---- ( -21.90, z-score = -2.16, R) >dp4.chr3 1314821 100 + 19779522 ACUUAGCAGCACUGUGUAUUUCACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCCGAGUGCACACACU-CAAACACACAUUUCCGCC--------------------- .....((((.((((((.....))))))...))))..(((((.......)))))(((((.....))))).(((((....))))-).................--------------------- ( -24.60, z-score = -3.40, R) >droPer1.super_2 1489204 100 + 9036312 ACUUAGCAGCACUGCGUAUUUCACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCCGAGUGCACACACU-CAAACACACAUUUCCGCC--------------------- .....((((.((((.(.....).))))...))))..(((((.......)))))(((((.....))))).(((((....))))-).................--------------------- ( -20.70, z-score = -2.31, R) >droWil1.scaffold_180700 2634256 115 + 6630534 ACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAAUGCACACACA-CACUGGCUCACACACACACACAUACGGUUUUUCCUUCU- .....(.-----((.((......((((....((((((((((.......))))((((((.....)))))).))))))......-.))))......)).)))........((.....))....- ( -18.40, z-score = -0.71, R) >droVir3.scaffold_10324 1198714 112 + 1288806 ACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCUUGAGUGCGCCUGCCGCCCACCCGCUAGCCCCCCGCGUCGAACGGAGGUU----- .....((-----.((((((((..(((....((((.((((((.......)))))))))).....)))...))))))))..)).((......)).((((..(((.......))).))))----- ( -24.50, z-score = 0.73, R) >droMoj3.scaffold_6496 22021915 113 - 26866924 ACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCUUGAGUGCGCAAACC--CCGGGCACCGACAUCUCGC--CAAGCCGCCCUCCCCCU .......-----(((((((((..(((....((((.((((((.......)))))))))).....)))...)))))))))....--..((((...((....))((--...)).))))....... ( -24.90, z-score = -0.79, R) >droGri2.scaffold_15245 9281271 107 + 18325388 ACUUAGC-----UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCUUGAGUGCGCACGCC-CCCUCCCCCUA----CACGCUAUCAGAGGAGGUU----- .....((-----(((((((((..(((....((((.((((((.......)))))))))).....)))...))))))))).)).-.(((((..((.----.........)).)))))..----- ( -26.70, z-score = -1.66, R) >consensus ACUUAGC_____UGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACACU_CAAAGGCACUCGCACACAC___CAUUCCAUUUCCUCC__ ...............................((((((((((.......))))((((((.....)))))).)))))).............................................. (-13.16 = -12.88 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:05:03 2011