
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,006,963 – 3,007,063 |
| Length | 100 |
| Max. P | 0.699607 |

| Location | 3,006,963 – 3,007,063 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.63 |
| Shannon entropy | 0.53466 |
| G+C content | 0.41921 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -13.54 |
| Energy contribution | -13.77 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.699607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
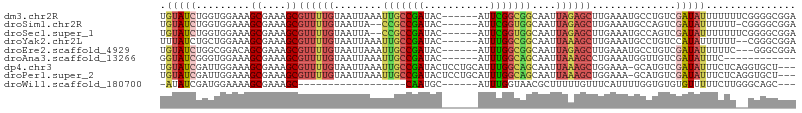
>dm3.chr2R 3006963 100 + 21146708 UGUAUCUGGUGGAAAGCGAAAGCGUUUUGUAAUUAAAUUGCCGAUAC------AUUCGGCGGCAAUUAGAGCUUGAAAUGCCUGUCGAUAUUUUUUUCGGGGCGGA .((.(((((..((((.(((..((((((((...((.((((((((...(------....).)))))))).))...))))))))...)))...))))..)))))))... ( -28.00, z-score = -0.99, R) >droSim1.chr2R 1806131 97 + 19596830 UGUAUCUGGUGGAAAGCGAAAGCGUUUUGUAAUUA--CCGCCGAUAC------AUUCGGUGGCAAUUAGAGCUUGAAAUGCCAGUCGAUAUUUUUU-CGGGGCGGA .((..(((((.....((....))((((..(((((.--(((((((...------..))))))).))))))))).......)))))((((.......)-))).))... ( -29.60, z-score = -1.55, R) >droSec1.super_1 676121 98 + 14215200 UGUAUCUGGUGGAAAGCGAAAGCGUUUUGUAAUUA--CCGCCGAUAC------AUUCGGUGGCAAUUAGAGCUUGAAAUGCCAGUCGAUAUUUUUUUCGGGGCGGA .((..(((((.....((....))((((..(((((.--(((((((...------..))))))).))))))))).......)))))((((........)))).))... ( -29.90, z-score = -1.61, R) >droYak2.chr2L 15718842 98 + 22324452 UUUAUCUGCUGGAAAGCGAAAGCGUUUUGUAAUUAAAUUGCCGAUAC------AUUUGGCGGCAAUUAAAGCUUGAAAUGCCUGUCCAUAUUUUUU--CGGGCGGA ....((((((.((((((....))((((((...((.((((((((...(------....).)))))))).))...))))))..............)))--).)))))) ( -29.30, z-score = -1.98, R) >droEre2.scaffold_4929 7080240 97 - 26641161 UGUAUCUGGCGGACAGCGAAAGCGUUUUGUAAUUAAAUUGCCGAUAC------AUUUGGCGGCAAUUAGAGCUUGAAAUGCCUGUCGAUAUUUUUC---GGGCGGA ....((((.(.(((((.....((((((((...((.((((((((...(------....).)))))))).))...)))))))))))))((......))---.).)))) ( -28.30, z-score = -1.11, R) >droAna3.scaffold_13266 3243959 88 + 19884421 GGUAUCGGGUGGAAAGCGAAAGCGUUUUGUAAUUAAAUUGCCGAUAC------AUUUGGCAGCAAUUAAAGCCUGAAAUGGUUGUCGAUAUUUC------------ (((((((((..(((.((....)).)))..).......(((((((...------..))))))).......((((......)))).))))))))..------------ ( -24.30, z-score = -2.25, R) >dp4.chr3 1295077 102 + 19779522 UGUAUCGAUUGGAAAGCGAAAGCGUUUUGUAAUUAAAUUGCCGAUACUCCUGCAUUUGGCAGCAAUUAAAGCUGGAAA-GCAUGUCGAUAUUUCUCAGGUGCU--- .((((((((((.((((((....)))))).))))).......(((((...((((.....))))........(((....)-)).)))))..........))))).--- ( -26.20, z-score = -0.79, R) >droPer1.super_2 1469405 102 + 9036312 UGUAUCGAUUGGAAAGCGAAAGCGUUUUGUAAUUAAAUUGCCGAUACUCCUGCAUUUGGCAGCAAUUAAAGCUGGAAA-GCAUGUCGAUAUUUCUCAGGUGCU--- .((((((((((.((((((....)))))).))))).......(((((...((((.....))))........(((....)-)).)))))..........))))).--- ( -26.20, z-score = -0.79, R) >droWil1.scaffold_180700 2604502 78 + 6630534 -AUAUCGAUGGAAAAGCGAAAGC------------------CAAUGC------AUUUGGUAACGCUUUUUGUUUCAUUUUGGUGUGUGUUUUUCUUGGGCAGC--- -(((((((((((((((((...((------------------(((...------..)))))..)))))....)))))))..))))).(((((.....)))))..--- ( -17.20, z-score = -0.23, R) >consensus UGUAUCUGGUGGAAAGCGAAAGCGUUUUGUAAUUAAAUUGCCGAUAC______AUUUGGCGGCAAUUAAAGCUUGAAAUGCCUGUCGAUAUUUUUU__GGGGCGGA .(((((.........((....))((((((........(((((((...........)))))))....))))))..............)))))............... (-13.54 = -13.77 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:59 2011