
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 3,000,575 – 3,000,683 |
| Length | 108 |
| Max. P | 0.860038 |

| Location | 3,000,575 – 3,000,683 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 68.60 |
| Shannon entropy | 0.56796 |
| G+C content | 0.58082 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -14.14 |
| Energy contribution | -15.75 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.550653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
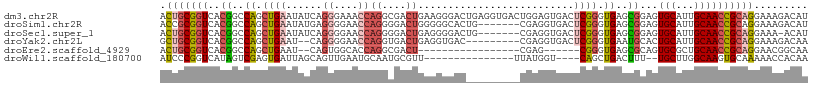
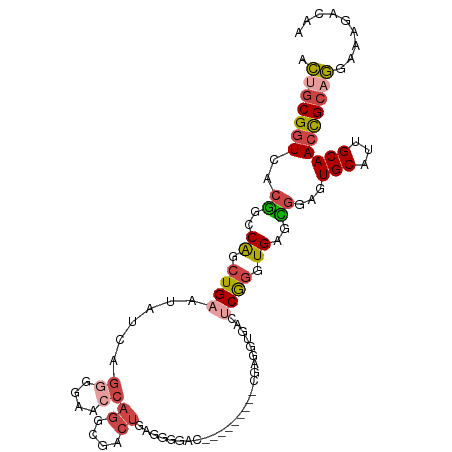
>dm3.chr2R 3000575 108 + 21146708 ACUGCGGUCACGGCCAGCUGAAUAUCAGGGAAACCAGGCGACUGAAGGGACUGAGGUGACUGGAGUGACUCGGGUGAGCGGAGUGCAUUGCAACCGCAGGAAAGACAU .((((((((((.(((...((.....))((....)).)))..((((.(..(((.((....)).).))..)))))))))((((......))))..))))))......... ( -36.50, z-score = -1.39, R) >droSim1.chr2R 1799487 101 + 19596830 ACCGCGGUCACGGCCAGCUGAAUAUGAGGGGAACCAGGGGACUGGGGGCACUG-------CGAGGUGACUCGGGUGAGCGGAGUGCAUUGCAACCGCAGGAAAGACAU .((((((((((..((.(((..............((((....))))...((((.-------((((....))))))))))))).))).......))))).))........ ( -38.21, z-score = -1.78, R) >droSec1.super_1 669765 100 + 14215200 ACUGCGGUCACGGCCAGCUGAAUAUCAGGGGAACCAGGGGACUGAGGGGACUG-------CGAGGUGACUCGGGUGAGCGGAGUGCAUUGCAACCGCAGGAAA-ACAU .((((((((((..((..(((.....))).))...(((....)))((....)).-------((((....)))).))))((((......))))..))))))....-.... ( -37.30, z-score = -2.19, R) >droYak2.chr2L 15712509 97 + 22324452 GCUGCGGUCACGGCCAGCUGAAU--CAGGGGAACCAGGUGACUGAGGUGAC---------CGAGGUGACUCGGGUGAAUGCACUGCAUUGCAACCGCAGGAAAGACAA .((((((((((.(((........--..((....))((....))..)))..(---------((((....))))))))).((((......)))).))))))......... ( -40.00, z-score = -2.66, R) >droEre2.scaffold_4929 7074395 83 - 26641161 ACUGCGGUCACGGCCAGCUGAAU--CAGUGGCACCAGGCGACU-----------------CGAG------CGGGUGAGCGCAGUGCGCUGCAACCGCAGGAACGGCAA .(((((((..((((..((((...--)))).((((...(((.((-----------------((..------....))))))).))))))))..)))))))......... ( -34.80, z-score = -0.28, R) >droWil1.scaffold_180700 2597327 87 + 6630534 AUCCCGGUCAUAGUCGAGUGAUUAGCAGUUGAAUGCAAUGCGUU---------------UUAUGGU----CAGCUGACUUU--UGCUUGGCAAGUGCAAAAACCACAA .....((((((.((((((..(..(((((((((((((...)))).---------------......)----)))))).)).)--..))))))..))).....))).... ( -21.71, z-score = -0.23, R) >consensus ACUGCGGUCACGGCCAGCUGAAUAUCAGGGGAACCAGGCGACUGAGGGGAC_________CGAGGUGACUCGGGUGAGCGGAGUGCAUUGCAACCGCAGGAAAGACAA .(((((((..((..((.((((......((....))((....))..........................)))).))..))...(((...))))))))))......... (-14.14 = -15.75 + 1.61)

| Location | 3,000,575 – 3,000,683 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 68.60 |
| Shannon entropy | 0.56796 |
| G+C content | 0.58082 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -14.44 |
| Energy contribution | -16.13 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
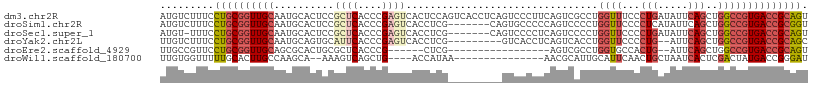
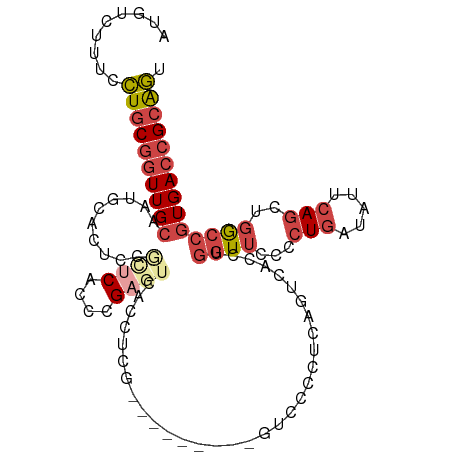
>dm3.chr2R 3000575 108 - 21146708 AUGUCUUUCCUGCGGUUGCAAUGCACUCCGCUCACCCGAGUCACUCCAGUCACCUCAGUCCCUUCAGUCGCCUGGUUUCCCUGAUAUUCAGCUGGCCGUGACCGCAGU .........(((((((..(..........((((....))))....(((((....((((.....((((....)))).....))))......)))))..)..))))))). ( -31.30, z-score = -2.00, R) >droSim1.chr2R 1799487 101 - 19596830 AUGUCUUUCCUGCGGUUGCAAUGCACUCCGCUCACCCGAGUCACCUCG-------CAGUGCCCCCAGUCCCCUGGUUCCCCUCAUAUUCAGCUGGCCGUGACCGCGGU .........(((((((..(...(((((.(((((....)))......))-------.)))))..((((....((((............))))))))..)..))))))). ( -29.60, z-score = -1.43, R) >droSec1.super_1 669765 100 - 14215200 AUGU-UUUCCUGCGGUUGCAAUGCACUCCGCUCACCCGAGUCACCUCG-------CAGUCCCCUCAGUCCCCUGGUUCCCCUGAUAUUCAGCUGGCCGUGACCGCAGU ....-....(((((((..(..(((.....((((....))))......)-------))................((((...(((.....)))..)))))..))))))). ( -28.60, z-score = -1.88, R) >droYak2.chr2L 15712509 97 - 22324452 UUGUCUUUCCUGCGGUUGCAAUGCAGUGCAUUCACCCGAGUCACCUCG---------GUCACCUCAGUCACCUGGUUCCCCUG--AUUCAGCUGGCCGUGACCGCAGC .........(((((((..(......(((.((((....)))))))...(---------((((.((.(((((...((....))))--))).)).))))))..))))))). ( -33.90, z-score = -2.30, R) >droEre2.scaffold_4929 7074395 83 + 26641161 UUGCCGUUCCUGCGGUUGCAGCGCACUGCGCUCACCCG------CUCG-----------------AGUCGCCUGGUGCCACUG--AUUCAGCUGGCCGUGACCGCAGU .........(((((((..(.(((((((((((((.....------...)-----------------)).)))..)))))(((((--...))).)))).)..))))))). ( -34.40, z-score = -0.97, R) >droWil1.scaffold_180700 2597327 87 - 6630534 UUGUGGUUUUUGCACUUGCCAAGCA--AAAGUCAGCUG----ACCAUAA---------------AACGCAUUGCAUUCAACUGCUAAUCACUCGACUAUGACCGGGAU ((((((((.(((.((((((...)))--..))))))..)----)))))))---------------...((((((....))).)))......((((........)))).. ( -17.00, z-score = 0.22, R) >consensus AUGUCUUUCCUGCGGUUGCAAUGCACUCCGCUCACCCGAGUCACCUCG_________GUCCCCUCAGUCACCUGGUUCCCCUGAUAUUCAGCUGGCCGUGACCGCAGU .........((((((((((..........((((....))))................................((((...(((.....)))..)))))))))))))). (-14.44 = -16.13 + 1.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:59 2011