
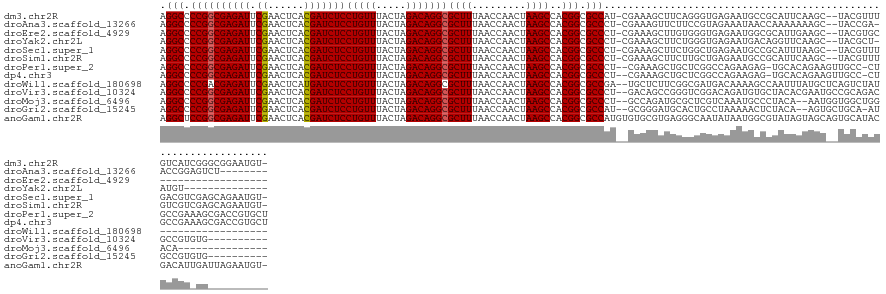
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,958,264 – 2,958,398 |
| Length | 134 |
| Max. P | 0.622348 |

| Location | 2,958,264 – 2,958,398 |
|---|---|
| Length | 134 |
| Sequences | 13 |
| Columns | 138 |
| Reading direction | forward |
| Mean pairwise identity | 71.74 |
| Shannon entropy | 0.63583 |
| G+C content | 0.54231 |
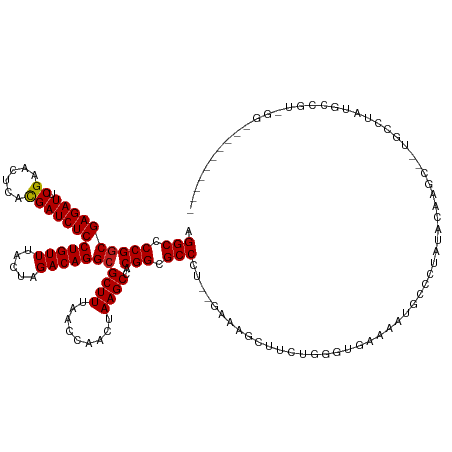
| Mean single sequence MFE | -36.44 |
| Consensus MFE | -23.99 |
| Energy contribution | -23.99 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.15 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2958264 134 + 21146708 AGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAU-CGAAAGCUUCAGGGUGAGAAUGCCGCAUUCAAGC--UACGUUUGUCAUCGGGCGGAAUGU- ..(((((((.(((((.((......)))))))))).......((((((((((.................))))))..-....(((((..(((((.(.....).)))))))))--).....))))...)))).......- ( -38.03, z-score = 0.56, R) >droAna3.scaffold_13266 3197424 126 + 19884421 AGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCU-CGAAAGUUCUUCCGUAGAAAUAACCAAAAAAAGC--UACCGA-ACCGGAGUCU-------- ((((.((((........(((((..(((.....((((.....))))((((((.................)))))).)-))..)))))(((.((((................)--))).))-))))).))))-------- ( -30.92, z-score = -0.64, R) >droEre2.scaffold_4929 7031499 117 - 26641161 AGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCU-CGAAAGCUUGUGGGUGAGAAUGGCGCAUUGAAGC--UACGUGC------------------ .(((....(((..((((..((((((((...((((((.....)))))).(((((..........((....)).....-..)))))))))))))..))))..)))......))--)......------------------ ( -35.03, z-score = 0.06, R) >droYak2.chr2L 15671295 120 + 22324452 AGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCU-CGAAAGCUUCUGGGUGAGAAUGACAGGUUCAAGC--UACGCU-AUGU-------------- .(((....(((((((.((......)))))))(((((.....)))))))((((.((((......((....))((((.-(....).....))))..........)))).))))--...)))-....-------------- ( -33.20, z-score = 0.24, R) >droSec1.super_1 626959 134 + 14215200 AGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCU-CGAAAGCUUCUGGCUGAGAAUGCCGCAUUUAAGC--UACGUUUGACGUCGAGCAGAAUGU- .(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))((-(((.(((((.((((.......)))).....))))--).((.....)))))))........- ( -37.80, z-score = -0.04, R) >droSim1.chr2R 1758377 134 + 19596830 AGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCU-CGAAAGCUUCUUGCUGAGAAUGCCGCAUUCAAGC--UACGUUUGUCGUCGAGCAGAAUGU- ..((....))(((((.((......)))))))((((((....(((((((((((.........))))..(((((..((-(...(((.....))))))..))))).........--...)))))))...)))))).....- ( -38.00, z-score = -0.17, R) >droPer1.super_2 1417226 134 + 9036312 AGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCU--CGAAAGCUGCUCGGCCAGAAGAG-UGCACAGAAGUUGCC-CUGCCGAAAGCGACCGUGCU .((....((((((((.((......))))))((((((.....))))))))))....)).....(((.(((((((..(--((...((.((((........)))-))).(((........-))).)))..))).))))))) ( -39.80, z-score = 0.23, R) >dp4.chr3 1241641 134 + 19779522 AGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCU--CGAAAGCUGCUCGGCCAGAAGAG-UGCACAGAAGUUGCC-CUGCCGAAAGCGACCGUGCU .((....((((((((.((......))))))((((((.....))))))))))....)).....(((.(((((((..(--((...((.((((........)))-))).(((........-))).)))..))).))))))) ( -39.80, z-score = 0.23, R) >droWil1.scaffold_180698 9356226 118 - 11422946 AGGCCCCGACGAGAUUCGAACUCAUGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCGA--UGCUCUUCGGCGAUGACAAAAGCCAAUUUAUGCUCAGUCUAU------------------ .(((.(((..((((((.........))))))(((((.....)))))(.((((.........))))).)))((((((--......)))))).........)))..................------------------ ( -29.20, z-score = -0.58, R) >droVir3.scaffold_10324 1108996 126 + 1288806 AGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCU--GACAGCCGGGUCGGACAGAUGUGCUACACGAAUGCCGCAGACGCCGUGUG---------- .((((((((((((((.((......)))))))............((((((((.................)))).)))--)...)))))).(((.((..((((...))))..))))).....))).....---------- ( -40.83, z-score = -0.55, R) >droMoj3.scaffold_6496 21932615 119 - 26866924 AGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCU--GCCAGAUGCGCUCGUCAAAUGCCCUACA--AAUGGUGGCUGGACA--------------- .(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))..--.(((((((....))))....((((((..--..))).))))))...--------------- ( -33.80, z-score = 0.05, R) >droGri2.scaffold_15245 9193676 123 + 18325388 AGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAU--GCGGGAUGCACUGCCUAAAAACUCUACA--AGUGCUGCA-AUGCCGUGUG---------- .(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))((--((((.(((((((................--)))))....-)).)))))).---------- ( -37.19, z-score = -0.99, R) >anoGam1.chr2R 51288761 137 - 62725911 AGGCUCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUGUGUGCGUGAGGGCAAUAUAAUGGCGUAUAGUAGCAGUGCAUACGACAUUGAUUAGAAUGU- .(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))...(((.(....).)))...(((((.(((((.(((....)))))))).)))))..........- ( -40.10, z-score = -0.34, R) >consensus AGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCU__GAAAGCUUCUGGGUGAAAAUGCCCUAUACAAGC__UGCCUAUGCCGU_GG__________ .(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))................................................................ (-23.99 = -23.99 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:56 2011