| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,957,469 – 2,957,564 |
| Length | 95 |
| Max. P | 0.562894 |

| Location | 2,957,469 – 2,957,564 |
|---|---|
| Length | 95 |
| Sequences | 14 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.94 |
| Shannon entropy | 0.44231 |
| G+C content | 0.53656 |
| Mean single sequence MFE | -27.31 |
| Consensus MFE | -17.79 |
| Energy contribution | -18.36 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.562894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
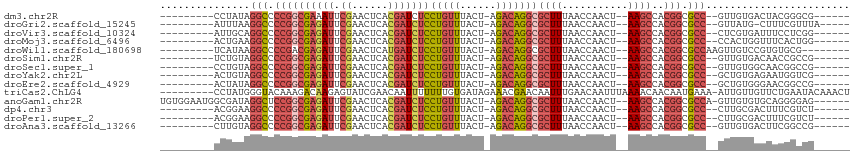
>dm3.chr2R 2957469 95 + 21146708 ---------CCUAUAGGCCCCGGCGAAAUUCGAACUCACGAUCUCCUGUUUACU-AGACAGGCGCUUUAACCAACU--AAGCCACGGCGCC--GUUGUGACUACGGGCG------ ---------.......((((.((((....(((......)))...((((((....-.))))))))))....(((((.--..((....))...--)))).).....)))).------ ( -26.10, z-score = -0.60, R) >droGri2.scaffold_15245 9192604 95 + 18325388 ---------AUUUAAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACU-AGACAGGCGCUUUAACCAACU--AAGCCACGGCGCC--GUUAUG-CUUUCGUUUA----- ---------......(((.((((((((((.((......)))))))(((((....-.)))))))((((.........--))))..))).)))--......-..........----- ( -25.20, z-score = -1.27, R) >droVir3.scaffold_10324 1107853 95 + 1288806 ---------AUUGCAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACU-AGACAGGCGCUUUAACCAACU--AAGCCACGGCGCC--CUCGUGAUUUCCUCGG------ ---------((..(.(((.((((((((((.((......)))))))(((((....-.)))))))((((.........--))))..))).)))--...)..))........------ ( -25.20, z-score = -0.65, R) >droMoj3.scaffold_6496 21931481 95 - 26866924 ---------ACUGAAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACU-AGACAGGCGCUUUAACCAACU--AAGCCACGGCGCC--CCACUGGUUUCACUGG------ ---------......(((.((((((((((.((......)))))))(((((....-.)))))))((((.........--))))..))).)))--(((.((....)).)))------ ( -26.00, z-score = -1.04, R) >droWil1.scaffold_180698 9354090 95 - 11422946 ---------UCAUAAGGCCCCGACGAGAUUCGAACUCAUGAUCUCCUGUUUACU-AGACAGGCGCUUUAACCAACU--AAGCCACGGCGCCAAGUUGUCCGUGUGCG-------- ---------...((((((.((...((((((.........)))))).((((....-.)))))).)))))).......--..(((((((((......)).))))).)).-------- ( -24.60, z-score = -1.00, R) >droSim1.chr2R 1757576 95 + 19596830 ---------UCUGUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACU-AGACAGGCGCUUUAACCAACU--AAGCCACGGCGCC--GUUGUGACAACCGCCG------ ---------......(((.((((((((((.((......)))))))(((((....-.)))))))((((.........--))))..))).)))--((((...)))).....------ ( -25.80, z-score = -0.56, R) >droSec1.super_1 626149 95 + 14215200 ---------CCUGUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACU-AGACAGGCGCUUUAACCAACU--AAGCCACGGCGCC--GUUGUGGCAACGGCCG------ ---------......((((..((((((((.((......))))))((((((....-.))))))))))..........--..((((((((...--))))))))...)))).------ ( -33.30, z-score = -1.82, R) >droYak2.chr2L 15670603 95 + 22324452 ---------ACUGUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACU-AGACAGGCGCUUUAACCAACU--AAGCCACGGCGCC--GCUGUGAGAAUGGUCG------ ---------......((((.(((.(((((.((......))))))))))(((((.-((...((((((..........--.......))))))--.)))))))...)))).------ ( -27.13, z-score = -0.38, R) >droEre2.scaffold_4929 7030700 95 - 26641161 ---------ACUAUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACU-AGACAGGCGCUUUAACCAACU--AAGCCACGGCGCC--GCUGUGGGAACGGCCG------ ---------......((((..((((((((.((......))))))((((((....-.))))))))))..........--...(((((((...--)))))))....)))).------ ( -32.00, z-score = -1.73, R) >triCas2.ChLG4 1555562 105 - 13894384 ---------CCUAUGGGUACAAAGACAAGAGUAUCGAACAAUUUUUUUGUGAUAGAAACGAACAAUUUGAACAAUUUAAAACAACAAUGAAA-AUUGUUGUUCUGAAUACAAACU ---------.......((.((((.......((.((..((((.....))))....)).))......)))).)).(((((.(((((((((....-))))))))).)))))....... ( -19.82, z-score = -1.97, R) >anoGam1.chr2R 51286906 105 - 62725911 UGUGGAAUGGCGAUAGGCUCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACU-AGACAGGCGCUUUAACCAACU--AAGCCACGGCGCCA-GUUGUGUGCAGGGGAG------ .((((.....(((....(((....)))..)))...))))..((((((((.(((.-.(((.((((((..........--.......)))))).-)))))).)))))))).------ ( -32.13, z-score = 0.06, R) >dp4.chr3 1240873 95 + 19779522 ---------ACGGAAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACU-AGACAGGCGCUUUAACCAACU--AAGCCACGGCGCC--CUUGCGACUUUCGUCU------ ---------((((((((((..((((((((.((......)))))))(((((....-.)))))...............--..)))..)))((.--...))..)))))))..------ ( -28.50, z-score = -1.44, R) >droPer1.super_2 1416457 95 + 9036312 ---------ACGGAAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACU-AGACAGGCGCUUUAACCAACU--AAGCCACGGCGCC--CUUGCGACUUUCGUCU------ ---------((((((((((..((((((((.((......)))))))(((((....-.)))))...............--..)))..)))((.--...))..)))))))..------ ( -28.50, z-score = -1.44, R) >droAna3.scaffold_13266 3196645 95 + 19884421 ---------CUUGUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACU-AGACAGGCGCUUUAACCAACU--AAGCCACGGCGCC--GUUGUGACUUCGGCCG------ ---------......((((.(((.(((((.((......)))))))))).((((.-.(((.((((((..........--.......))))))--)))))))....)))).------ ( -28.13, z-score = -0.84, R) >consensus _________ACUGUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACU_AGACAGGCGCUUUAACCAACU__AAGCCACGGCGCC__GUUGUGACUACGGCCG______ ...............(((.((((((((((.((......)))))))(((((......)))))))((((...........))))..))).)))........................ (-17.79 = -18.36 + 0.57)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:55 2011