
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,262,880 – 2,262,978 |
| Length | 98 |
| Max. P | 0.998961 |

| Location | 2,262,880 – 2,262,978 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 83.77 |
| Shannon entropy | 0.27241 |
| G+C content | 0.51130 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -27.03 |
| Energy contribution | -28.15 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.997386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2262880 98 + 23011544 -----ACCCGCAGCGCAGACACACACUCACACAC----UUAAAUAAUUUAAUAUUCCGAGUGUUUCGACAUCGAAAUUUGCACUUCCAUGG-----GCGGCGCUGCGGGCGA -----.((((((((((......((((((......----((((.....))))......))))))((((....))))...(((.((.....))-----)))))))))))))... ( -33.50, z-score = -4.00, R) >droYak2.chr2L 2244157 112 + 22324452 UCCCCCUCCCCUACACACACACACAAGCACAUACACGCUUAAAUAAUUUAAUAUUCAGAGUGUUUCGACAUCGAAAUUUGCACUUCGAUGGCCUGCGCUGCGUUGCGGGCGU ....................((((((((........)))).((((......))))....)))).....(((((((........)))))))(((((((......))))))).. ( -24.10, z-score = -0.85, R) >droEre2.scaffold_4929 2300109 103 + 26641161 ----CCCCCGCAGCACACACACACACACACACUCACGCUUAAAUAAUUUAAUAUUCAGAGUGUUUCGACAUCGAAAUUUGCACUUCGAUGG-----CCUGCGCUGCGGGCGU ----..((((((((.((..(........((((((.......................)))))).....(((((((........))))))))-----..)).))))))))... ( -31.70, z-score = -4.09, R) >droSec1.super_5 437887 101 + 5866729 CCGCACCCCGCAGCGCAGACACACACUCACGC------UUAAAUAAUUUAAUAUUCCGAGUGUUUCGACAUCGAAAUUUGCACUUCGAUGG-----GCGGCGCUGCGGGCGU ....((((((((((((......((((((....------...................))))))..((.(((((((........))))))).-----.)))))))))))).)) ( -39.00, z-score = -3.84, R) >droSim1.chr2L 2221657 101 + 22036055 CCGCCCCCCGCAGCGCAGACACACACUCACGC------UUAAAUAAUUUAAUAUUCCGAGUGUUUCGACAUCGAAAUUUGCACUUCGAUGG-----GCGGCGCUGCGGGCGU .(((((...(((((((......((((((....------...................))))))..((.(((((((........))))))).-----.)))))))))))))). ( -38.60, z-score = -3.49, R) >consensus _C_CCCCCCGCAGCGCAGACACACACUCACAC_C____UUAAAUAAUUUAAUAUUCCGAGUGUUUCGACAUCGAAAUUUGCACUUCGAUGG_____GCGGCGCUGCGGGCGU ......((((((((((......((((((.............................)))))).....(((((((........))))))).........))))))))))... (-27.03 = -28.15 + 1.12)

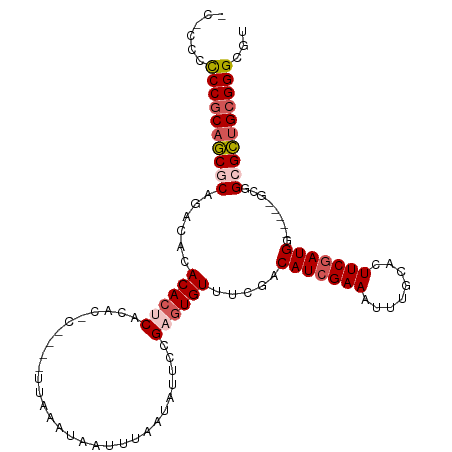
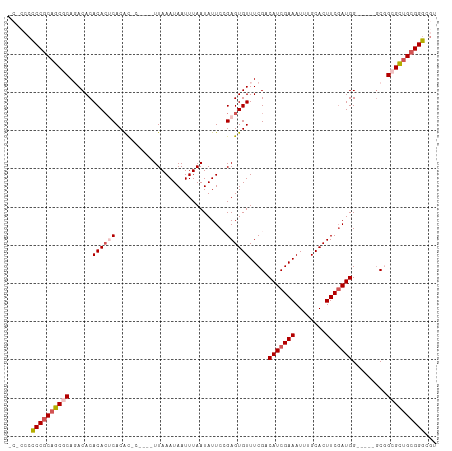
| Location | 2,262,880 – 2,262,978 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.77 |
| Shannon entropy | 0.27241 |
| G+C content | 0.51130 |
| Mean single sequence MFE | -43.28 |
| Consensus MFE | -31.88 |
| Energy contribution | -32.72 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.998961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2262880 98 - 23011544 UCGCCCGCAGCGCCGC-----CCAUGGAAGUGCAAAUUUCGAUGUCGAAACACUCGGAAUAUUAAAUUAUUUAA----GUGUGUGAGUGUGUGUCUGCGCUGCGGGU----- ..(((((((((((...-----.(((.(((((....))))).)))..((.(((((((.(...((((.....))))----...).)))))))...)).)))))))))))----- ( -41.90, z-score = -4.70, R) >droYak2.chr2L 2244157 112 - 22324452 ACGCCCGCAACGCAGCGCAGGCCAUCGAAGUGCAAAUUUCGAUGUCGAAACACUCUGAAUAUUAAAUUAUUUAAGCGUGUAUGUGCUUGUGUGUGUGUGUAGGGGAGGGGGA ...(((...((((((((((((((((((((((....))))))))).....((((.(((((((......))))).)).))))....)))))))).)))))...)))........ ( -40.00, z-score = -2.46, R) >droEre2.scaffold_4929 2300109 103 - 26641161 ACGCCCGCAGCGCAGG-----CCAUCGAAGUGCAAAUUUCGAUGUCGAAACACUCUGAAUAUUAAAUUAUUUAAGCGUGAGUGUGUGUGUGUGUGUGUGCUGCGGGGG---- .(.(((((((((((.(-----((((((((((....))))))))).((..((((((((((((......)))))).....))))))...))...)).))))))))))).)---- ( -45.70, z-score = -5.10, R) >droSec1.super_5 437887 101 - 5866729 ACGCCCGCAGCGCCGC-----CCAUCGAAGUGCAAAUUUCGAUGUCGAAACACUCGGAAUAUUAAAUUAUUUAA------GCGUGAGUGUGUGUCUGCGCUGCGGGGUGCGG ((.((((((((((...-----.(((((((((....)))))))))..((.((((((((((((......)))))..------...)))))))...)).)))))))))))).... ( -43.60, z-score = -3.69, R) >droSim1.chr2L 2221657 101 - 22036055 ACGCCCGCAGCGCCGC-----CCAUCGAAGUGCAAAUUUCGAUGUCGAAACACUCGGAAUAUUAAAUUAUUUAA------GCGUGAGUGUGUGUCUGCGCUGCGGGGGGCGG .(.((((((((((...-----.(((((((((....)))))))))..((.((((((((((((......)))))..------...)))))))...)).)))))))))).).... ( -45.20, z-score = -3.81, R) >consensus ACGCCCGCAGCGCCGC_____CCAUCGAAGUGCAAAUUUCGAUGUCGAAACACUCGGAAUAUUAAAUUAUUUAA____G_GUGUGAGUGUGUGUCUGCGCUGCGGGGGG_G_ ...((((((((((.........(((((((((....))))))))).....((((((.(((((......)))))............))))))......))))))))))...... (-31.88 = -32.72 + 0.84)

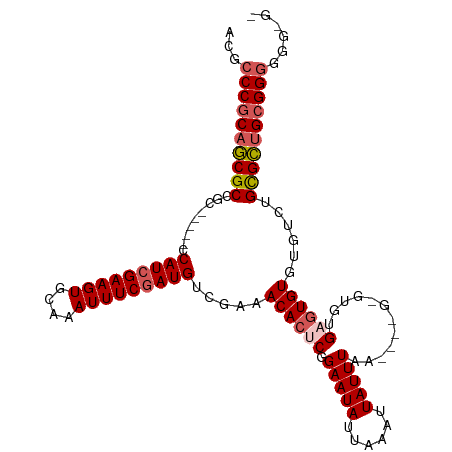
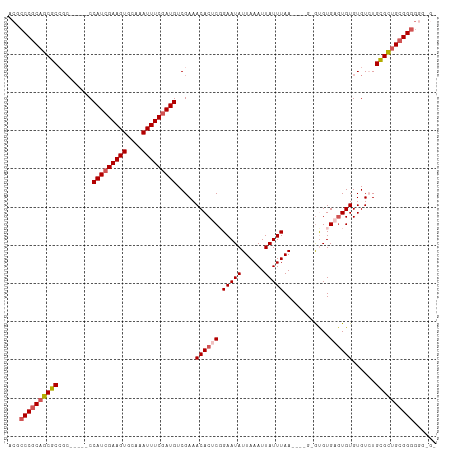
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:38 2011