
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,926,394 – 2,926,553 |
| Length | 159 |
| Max. P | 0.799317 |

| Location | 2,926,394 – 2,926,553 |
|---|---|
| Length | 159 |
| Sequences | 3 |
| Columns | 171 |
| Reading direction | forward |
| Mean pairwise identity | 70.72 |
| Shannon entropy | 0.39835 |
| G+C content | 0.40012 |
| Mean single sequence MFE | -45.87 |
| Consensus MFE | -19.38 |
| Energy contribution | -19.73 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.799317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
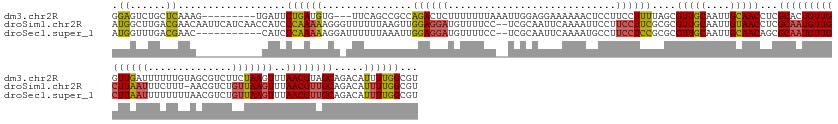
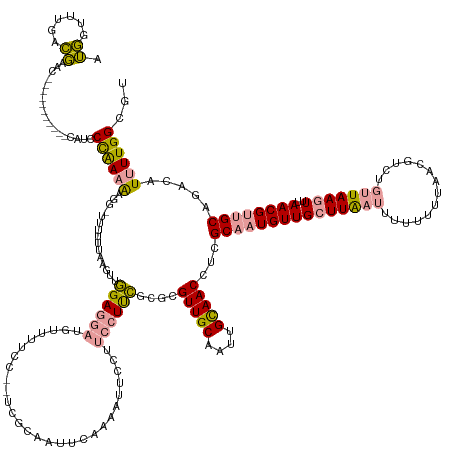
>dm3.chr2R 2926394 159 + 21146708 GGAGUCUGCUCAAAG---------UGAUUCUGAUGUG---UUCAGCCGCCAGACUCUUUUUUUAAAUUGGAGGAAAAAACUCCUUCCUUUUAGCGUUGCAAUUGCAACCUCGCACUGUUGGUUGAUUUUUUGUAGCGUCUUCUAAGUUUAACGUAGCAGACAUUUUGGCGU ((((((((......(---------((...((((....---.)))).))))))))))).....((((..((((((......))))))..))))(((((((((...(((((..........))))).....)))))))((((.(((.(.....).))).))))......)).. ( -44.20, z-score = -1.40, R) >droSim1.chr2R 1726598 168 + 19596830 AUGGCUUGACGAACAAUUCAUCAACCAUCCCAAAAAGGGUUUUUUAAGUUGGAGGAUGUUUUCC--UCGCAAUUCAAAAUUCCUUCCUUCGCGCGUUGCAAUUGUAACCUCGCAAUGUUGCUUAAUUUCUUU-AACGUCUGUUAAGUUUAACGUUGCAGACAUUUUGGCGU ((((.((((.((.....)).)))))))).......((((..((((.((((((((((.....)))--)).))))).))))..)))).....(((((((((....)))))...(((((((((((((((..(...-...)...)))))))..))))))))..........)))) ( -46.10, z-score = -2.98, R) >droSec1.super_1 594073 158 + 14215200 AUGGUUUGACGAAC-----------CAUCCCAAAAAGGAUUUUUUAAAUUGGAGGAUGUUUUCC--UCGCAAUUCAAAAUGCCUUCCUCCGCGCGUUGCAAUUGCAACAGCGCAAUGUUGCUUAAUUUUUUUUAACGUCUGUUAAGUUUAACGUUGCAGACAUUUUGGCGU (((((((...))))-----------))).(((((((((...((((.((((((((((.....)))--)).))))).))))..)))......(((((((((....))))).)))).......................((((((...(.....)...)))))).))))))... ( -47.30, z-score = -3.09, R) >consensus AUGGUUUGACGAAC___________CAUCCCAAAAAGG_UUUUUUAAGUUGGAGGAUGUUUUCC__UCGCAAUUCAAAAUUCCUUCCUUCGCGCGUUGCAAUUGCAACCUCGCAAUGUUGCUUAAUUUUUUUUAACGUCUGUUAAGUUUAACGUUGCAGACAUUUUGGCGU .((......))..................((((((...............((((((............................))))))....(((((....)))))...(((((((((((((((..............)))))))..)))))))).....))))))... (-19.38 = -19.73 + 0.35)
| Location | 2,926,394 – 2,926,553 |
|---|---|
| Length | 159 |
| Sequences | 3 |
| Columns | 171 |
| Reading direction | reverse |
| Mean pairwise identity | 70.72 |
| Shannon entropy | 0.39835 |
| G+C content | 0.40012 |
| Mean single sequence MFE | -44.78 |
| Consensus MFE | -15.96 |
| Energy contribution | -15.75 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
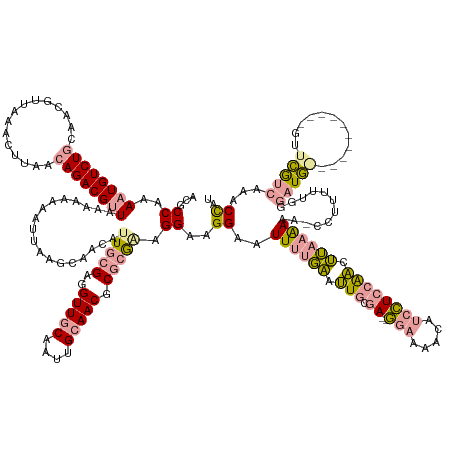
>dm3.chr2R 2926394 159 - 21146708 ACGCCAAAAUGUCUGCUACGUUAAACUUAGAAGACGCUACAAAAAAUCAACCAACAGUGCGAGGUUGCAAUUGCAACGCUAAAAGGAAGGAGUUUUUUCCUCCAAUUUAAAAAAAGAGUCUGGCGGCUGAA---CACAUCAGAAUCA---------CUUUGAGCAGACUCC ..........(((((((.(((((.((((......(((.((................)))))(((((((....))))).))....(((.((((....)))))))............)))).))))).((((.---....)))).....---------.....)))))))... ( -38.79, z-score = -1.36, R) >droSim1.chr2R 1726598 168 - 19596830 ACGCCAAAAUGUCUGCAACGUUAAACUUAACAGACGUU-AAAGAAAUUAAGCAACAUUGCGAGGUUACAAUUGCAACGCGCGAAGGAAGGAAUUUUGAAUUGCGA--GGAAAACAUCCUCCAACUUAAAAAACCCUUUUUGGGAUGGUUGAUGAAUUGUUCGUCAAGCCAU .(((...(((((.(((((((((..........))))))-.((....))..))))))))(((..((.......))..)))))).(((((((..((((((.(((.((--(((.....)))))))).))))))...)))))))...(((((((((((.....))))).)))))) ( -44.90, z-score = -2.30, R) >droSec1.super_1 594073 158 - 14215200 ACGCCAAAAUGUCUGCAACGUUAAACUUAACAGACGUUAAAAAAAAUUAAGCAACAUUGCGCUGUUGCAAUUGCAACGCGCGGAGGAAGGCAUUUUGAAUUGCGA--GGAAAACAUCCUCCAAUUUAAAAAAUCCUUUUUGGGAUG-----------GUUCGUCAAACCAU ..(((..((((((((...............))))))))................(.((((((.(((((....))))))))))).)...))).((((((((((.((--(((.....))))))))))))))).(((((....)))))(-----------(((.....)))).. ( -50.66, z-score = -4.31, R) >consensus ACGCCAAAAUGUCUGCAACGUUAAACUUAACAGACGUUAAAAAAAAUUAAGCAACAUUGCGAGGUUGCAAUUGCAACGCGCGAAGGAAGGAAUUUUGAAUUGCGA__GGAAAACAUCCUCCAACUUAAAAAA_CCUUUUUGGGAUG___________GUUCGUCAAACCAU ...((..((((((((...............))))))))..................(((((..(((((....))))).))))).))..((...(((((.........(((.....)))(((((...............)))))...................))))))).. (-15.96 = -15.75 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:53 2011