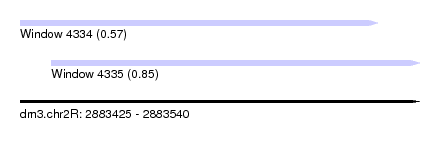
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,883,425 – 2,883,540 |
| Length | 115 |
| Max. P | 0.847868 |

| Location | 2,883,425 – 2,883,528 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 71.96 |
| Shannon entropy | 0.45353 |
| G+C content | 0.47242 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -15.47 |
| Energy contribution | -17.59 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.574853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
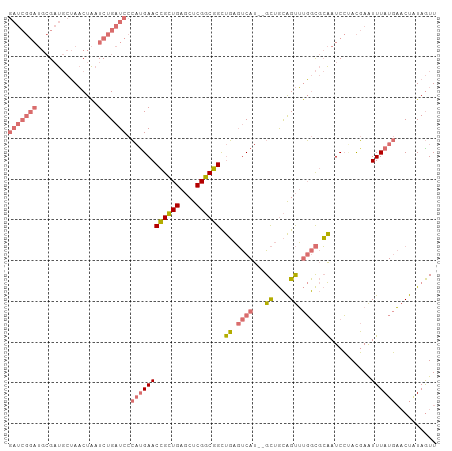
>dm3.chr2R 2883425 103 + 21146708 GAUCGGAUGCGAUGCUAACUAUUCUGAUCCCACGAACCGCUGAGCUCGGCGGCUGAGUCAU--GCUGCAGUUUGGCGCAAUCCUACGAAUUUAUGAACUAUAGUU (((((((...............)))))))........(((..((((..(((((........--)))))))))..)))............................ ( -25.36, z-score = 0.35, R) >droAna3.scaffold_13266 9193155 94 + 19884421 ----------GUUCCGAUCUGAUCUGCUCCAUUGAACUGAUGAACUCGUCAGUCUCUCCAUUAGUCAUGGCGCUAUUAGAUCGAAUUAGUUUAUCU-CUCUAAUU ----------....(((((((((..((.((((.(((((((((....)))))))...........)))))).)).)))))))))((((((.......-..)))))) ( -27.10, z-score = -3.77, R) >droSec1.super_1 555721 103 + 14215200 GAUCGGAUGCGAUGCUAACUAAUCUGAUCCCAUGAACCGCUGAGCUCGGCGGCUGAGUCAU--GCUGCAGUUUGGCGCAAUCCUACGAAUUUAUGAACUGUAGUU ((((((((.............)))))))).(((((.((((((....)))))).....))))--)((((((((((.((........))......)))))))))).. ( -31.62, z-score = -1.42, R) >droSim1.chr2R 1690474 103 + 19596830 GAUCGGAUGCGAUGCUAACUAAUCUGAUCCCAUGAACCGCUGAGCUCGGCGGCUGAGUCAU--GCUGCAGUUUGGCGCAAUCCCACGAAUUUAUGAACUAUAGUU ((((((((.............)))))))).((((((.(((..((((..(((((........--)))))))))..)))...((....)).)))))).......... ( -28.72, z-score = -0.63, R) >consensus GAUCGGAUGCGAUGCUAACUAAUCUGAUCCCAUGAACCGCUGAGCUCGGCGGCUGAGUCAU__GCUGCAGUUUGGCGCAAUCCUACGAAUUUAUGAACUAUAGUU ((((((((.............)))))))).((((((((((((....)))))).((.((((...((....)).)))).))..........)))))).......... (-15.47 = -17.59 + 2.12)


| Location | 2,883,434 – 2,883,540 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 68.69 |
| Shannon entropy | 0.50801 |
| G+C content | 0.47144 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -18.01 |
| Energy contribution | -17.32 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2883434 106 + 21146708 CGAUGCUAACUAUUCUGAUCCCACGAACCGCUGAGCUCGGCGGCUGAGUCAU--GCUGCAGUUUGGCGCAAUCCUACGAAUUUAUGAACUAUAGUUUGGCUGGGGGUG .................(((((.....((((((....))))))...(((((.--((((.((((((.((........))......)))))).)))).))))).))))). ( -29.10, z-score = 0.21, R) >droAna3.scaffold_13266 9193163 93 + 19884421 ---------CUGAUCUGCUCCAUUGAACUGAUGAACUCGUCAGUCUCUCCAUUAGUCAUGGCGCUAUUAGAUCGAAUUAGUUUAUCU-CUCUAAUUAAAUAGA----- ---------.((((((((.((((.(((((((((....)))))))...........)))))).))....))))))((((((.......-..)))))).......----- ( -20.40, z-score = -1.44, R) >droSec1.super_1 555730 106 + 14215200 CGAUGCUAACUAAUCUGAUCCCAUGAACCGCUGAGCUCGGCGGCUGAGUCAU--GCUGCAGUUUGGCGCAAUCCUACGAAUUUAUGAACUGUAGUUUGACAUGAGGAG .(((........)))...((((((...((((((....))))))....((((.--(((((((((((.((........))......))))))))))).))))))).))). ( -33.70, z-score = -2.15, R) >droSim1.chr2R 1690483 106 + 19596830 CGAUGCUAACUAAUCUGAUCCCAUGAACCGCUGAGCUCGGCGGCUGAGUCAU--GCUGCAGUUUGGCGCAAUCCCACGAAUUUAUGAACUAUAGUUUGACAUGGGGAG .(((........)))...(((((((...(((..((((..(((((........--)))))))))..)))........((((((..........)))))).))))))).. ( -31.90, z-score = -1.31, R) >consensus CGAUGCUAACUAAUCUGAUCCCAUGAACCGCUGAGCUCGGCGGCUGAGUCAU__GCUGCAGUUUGGCGCAAUCCUACGAAUUUAUGAACUAUAGUUUGACAGGGGGAG ..................(((((((..((((((....)))))).((.((((...((....)).)))).))......((((((..........)))))).))))))).. (-18.01 = -17.32 + -0.69)

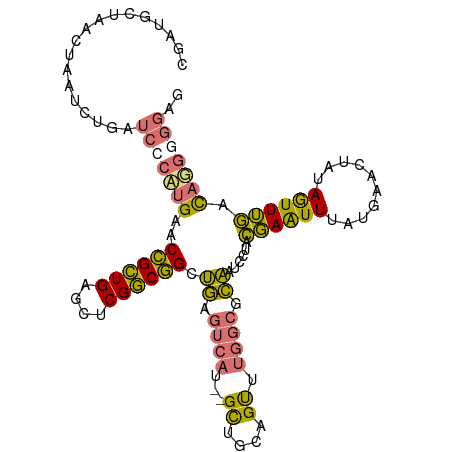

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:44 2011