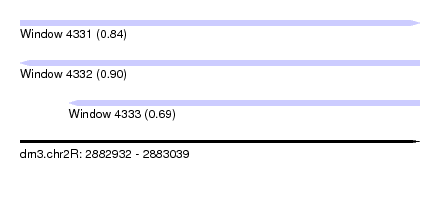
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,882,932 – 2,883,039 |
| Length | 107 |
| Max. P | 0.897133 |

| Location | 2,882,932 – 2,883,039 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.79 |
| Shannon entropy | 0.24157 |
| G+C content | 0.39672 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -19.54 |
| Energy contribution | -22.10 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.838942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2882932 107 + 21146708 AUUUUUUGUUGAUGG-GAAAUCGGUAAACGAGAUGGAU-----GACCUAUCUACUUGAAAUAGGUCAUUUGAUUUGUUCAAGAGUUCCAAUCCGCAGUUCUCCAACCUUUUUG .....((((.(((.(-(((.((....((((((.(..((-----((((((((.....))..))))))))..).))))))...)).)))).))).))))................ ( -27.60, z-score = -1.73, R) >droSim1.chr2R 1690010 98 + 19596830 -CUUUUUGUUGAUGGAGAAAUCGGUCAACGAGAUGGAUGGGAUGACCUAUCUGUUUGAAAUAGGUCAUUUGAUU--------------GCUCCGCAGUUCUGAAACCUUUUUG -..((((((((((.((....)).)))))))))).((...((((((((((((.....))..))))))))))((((--------------((...))))))......))...... ( -31.10, z-score = -3.15, R) >droSec1.super_1 555259 98 + 14215200 -CUUUUUGUUGAUGGGGAAAUCGGUCAACGAGAUGGAUGGGAUGACCUAUCUAUUUGAAAUAGGUCAUUUGAUU--------------GCUCCGCAGUUCUCAAACCUUUUUG -....((((((((.((....)).))))))))((((((((((....))))))))))..(((.((((.....((((--------------((...)))))).....)))).))). ( -28.50, z-score = -2.07, R) >consensus _CUUUUUGUUGAUGG_GAAAUCGGUCAACGAGAUGGAUGGGAUGACCUAUCUAUUUGAAAUAGGUCAUUUGAUU______________GCUCCGCAGUUCUCAAACCUUUUUG ...((((((((((.((....)).))))))))))..(((.((((((((((((.....))..)))))))))).)))....................................... (-19.54 = -22.10 + 2.56)

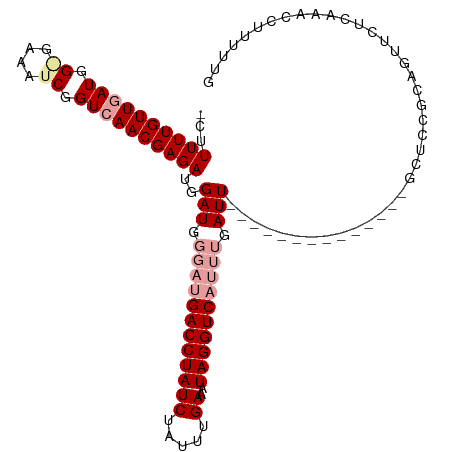
| Location | 2,882,932 – 2,883,039 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Shannon entropy | 0.24157 |
| G+C content | 0.39672 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -14.54 |
| Energy contribution | -15.87 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.897133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2882932 107 - 21146708 CAAAAAGGUUGGAGAACUGCGGAUUGGAACUCUUGAACAAAUCAAAUGACCUAUUUCAAGUAGAUAGGUC-----AUCCAUCUCGUUUACCGAUUUC-CCAUCAACAAAAAAU ......(((.(((((.(.((((..((((....((((.....)))).((((((((((.....)))))))))-----)))))..)))).....).))))-).))).......... ( -26.60, z-score = -2.46, R) >droSim1.chr2R 1690010 98 - 19596830 CAAAAAGGUUUCAGAACUGCGGAGC--------------AAUCAAAUGACCUAUUUCAAACAGAUAGGUCAUCCCAUCCAUCUCGUUGACCGAUUUCUCCAUCAACAAAAAG- ......((((....))))..((((.--------------((((..(((((((((((.....)))))))))))......((......))...)))).))))............- ( -22.20, z-score = -2.99, R) >droSec1.super_1 555259 98 - 14215200 CAAAAAGGUUUGAGAACUGCGGAGC--------------AAUCAAAUGACCUAUUUCAAAUAGAUAGGUCAUCCCAUCCAUCUCGUUGACCGAUUUCCCCAUCAACAAAAAG- ......(((((((((......((..--------------..))..(((((((((((.....)))))))))))........)))))..))))(((......))).........- ( -22.30, z-score = -2.61, R) >consensus CAAAAAGGUUUGAGAACUGCGGAGC______________AAUCAAAUGACCUAUUUCAAAUAGAUAGGUCAUCCCAUCCAUCUCGUUGACCGAUUUC_CCAUCAACAAAAAG_ ......((((.((((.....(((......................(((((((((((.....)))))))))))....))).))))...))))(((......))).......... (-14.54 = -15.87 + 1.33)



| Location | 2,882,945 – 2,883,039 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Shannon entropy | 0.25712 |
| G+C content | 0.42157 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -13.20 |
| Energy contribution | -14.87 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2882945 94 - 21146708 CAAAAAGGUUGGAGAACUGCGGAUUGGAACUCUUGAACAAAUCAAAUGACCUAUUUCAAGUAGAUAGGU-----CAUCCAUCUCGUUUACCGAUUUCCC- .....(((((((.((((...((((.(((....((((.....)))).((((((((((.....))))))))-----))))))))).)))).)))))))...- ( -25.60, z-score = -2.21, R) >droSim1.chr2R 1690022 86 - 19596830 CAAAAAGGUUUCAGAACUGCGGAGC--------------AAUCAAAUGACCUAUUUCAAACAGAUAGGUCAUCCCAUCCAUCUCGUUGACCGAUUUCUCC ......((((....))))..((((.--------------((((..(((((((((((.....)))))))))))......((......))...)))).)))) ( -20.50, z-score = -2.29, R) >droSec1.super_1 555271 86 - 14215200 CAAAAAGGUUUGAGAACUGCGGAGC--------------AAUCAAAUGACCUAUUUCAAAUAGAUAGGUCAUCCCAUCCAUCUCGUUGACCGAUUUCCCC ......(((((((((......((..--------------..))..(((((((((((.....)))))))))))........)))))..))))......... ( -22.10, z-score = -2.59, R) >consensus CAAAAAGGUUUGAGAACUGCGGAGC______________AAUCAAAUGACCUAUUUCAAAUAGAUAGGUCAUCCCAUCCAUCUCGUUGACCGAUUUCCCC ......((((.((((.....(((......................(((((((((((.....)))))))))))....))).))))...))))......... (-13.20 = -14.87 + 1.67)

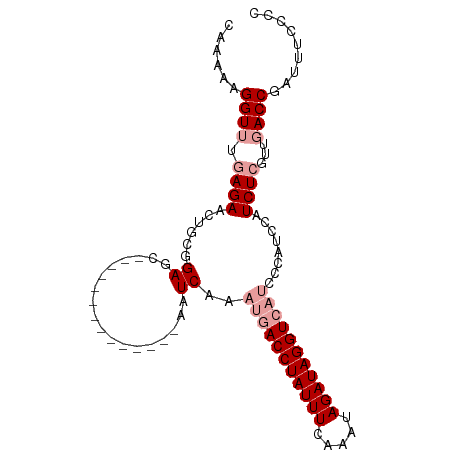
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:42 2011