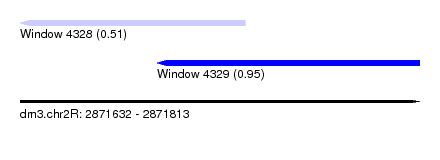
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,871,632 – 2,871,813 |
| Length | 181 |
| Max. P | 0.949894 |

| Location | 2,871,632 – 2,871,734 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.90 |
| Shannon entropy | 0.33385 |
| G+C content | 0.45438 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -15.34 |
| Energy contribution | -16.15 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.509005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
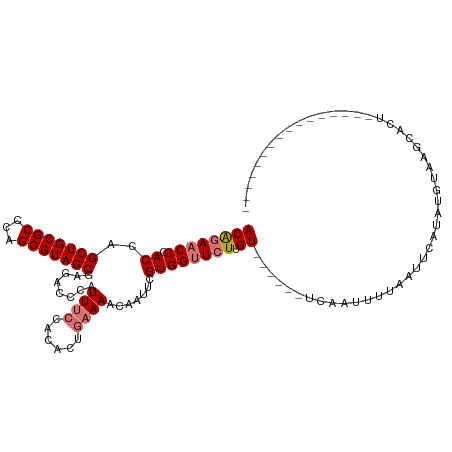
>dm3.chr2R 2871632 102 - 21146708 ACAGAACCCACCAGCUGCGCCCAGCGUAGCGAGACCCAUUUCCACACUGAAAACAAUUCGUGGUUCUUGU------UC-AUUUUAAUUCAUAUGCAAGCACUGGGAAAC--------- .............(((((((...)))))))....((((......(((.(((.....))))))((.(((((------..-..............))))).))))))....--------- ( -23.29, z-score = -1.20, R) >droEre2.scaffold_4929 6954128 118 + 26641161 ACGUAACCCACCAGCUGCGCCCAGCGUAGCGAGACCCAUUGGCACACUGAAAACAAUUGGUGGCUUUUGUAAUCAGUUUCUUUUAAAUCAGUUUUAUUCCCAUGAAAAUGCUUGCAAG .......(((((((((((((...)))))))..(.((....)))..............))))))...((((((.((.((((...((((.....)))).......)))).)).)))))). ( -26.80, z-score = -0.69, R) >droSim1.chr2R 1680287 96 - 19596830 ACAGAACCCACCAGCUGCGCCCAGCGUAGCGAAACCCAUUUCCACACUGAAAGCAAUUCGUGGUUCUUGU------UCAAUUUUAAUUCAUAUGUAAGCACU---------------- ((((((((.((..(((((((...)))))))(((.....((((......))))....)))))))))).)))------..........................---------------- ( -20.70, z-score = -1.96, R) >droSec1.super_1 545576 96 - 14215200 ACAGAACCCACCAGCUGCGCCCAGCGUAGCGAGACCCAUUUCCACACUGAAAGCAAUUCGUGGUUCUAGU------UCAAUUUUAAUUCAUAUGUAAGCACU---------------- ..((((((.((..(((((((...)))))))(((.....((((......))))....)))))))))))...------..........................---------------- ( -20.80, z-score = -1.98, R) >consensus ACAGAACCCACCAGCUGCGCCCAGCGUAGCGAGACCCAUUUCCACACUGAAAACAAUUCGUGGUUCUUGU______UCAAUUUUAAUUCAUAUGUAAGCACU________________ ..((((((.((..(((((((...)))))))........((((......)))).......))))))))................................................... (-15.34 = -16.15 + 0.81)

| Location | 2,871,694 – 2,871,813 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.62 |
| Shannon entropy | 0.14294 |
| G+C content | 0.59062 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -26.06 |
| Energy contribution | -26.54 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.949894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
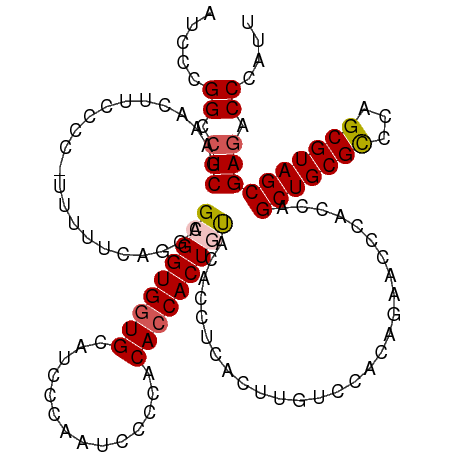
>dm3.chr2R 2871694 119 - 21146708 AUCCCGGCCUCGAAACUUCCCC-UUUUUCAGGAGGGGUGCUGCAUCCCAAUCCCCACACCACUGUACACCUCACUUGUCUACAGAACCCACCAGCUGCGCCCAGCGUAGCGAGACCCAUU .....((.(((.......((((-(((....))))))).(((((..................(((((.((.......)).))))).........((((....)))))))))))).)).... ( -32.80, z-score = -2.43, R) >droEre2.scaffold_4929 6954206 119 + 26641161 AUCCCGGCCUCAAAACUUCCCC-UUUCUCAGGGGCGGUGGUGCAUCCCAAUCCCCACACCACUCUGUACCUCACUUGUCCACGUAACCCACCAGCUGCGCCCAGCGUAGCGAGACCCAUU .....((.(((...(((.((((-(.....))))).)))((((((....................)))))).......................(((((((...)))))))))).)).... ( -32.05, z-score = -1.87, R) >droYak2.chr2L 15595516 120 - 22324452 AUCCCGGCCUCAAAACUUCCCCUUUUCUCUGGGGCGGUGGUGCAUCCCAAUCCCCACACCACUGUAUCCCUCACUUGUCCAUGUAACCCCCCAGCUGCGUCCAGCGUAGCGAGACCCAUU .....((.(((..................(((((((((((((..............))))))))........((........))....)))))((((((.....))))))))).)).... ( -33.84, z-score = -2.67, R) >droSim1.chr2R 1680343 119 - 19596830 AUCCCGGCCUCGAAACUUCCCC-UUUUCCAGCAGCGGUGGUGCAUCCCAAUCCCCACACCACUGUACACCUCACUUGUCCACAGAACCCACCAGCUGCGCCCAGCGUAGCGAAACCCAUU .....((..((((((.......-.)))).....(((((((((..............)))))))))..................))..))....(((((((...))))))).......... ( -26.84, z-score = -2.14, R) >droSec1.super_1 545632 119 - 14215200 AUCCCGGCCUCGAAACUUCCCC-UUUUCCAGCAGCGGUGGUGCAUCCCAAUCCCCACACCACUGCACACCUCACUUGUCCACAGAACCCACCAGCUGCGCCCAGCGUAGCGAGACCCAUU .....((.(((...........-..........(((((((((..............)))))))))............................(((((((...)))))))))).)).... ( -31.84, z-score = -3.25, R) >consensus AUCCCGGCCUCGAAACUUCCCC_UUUUUCAGGAGCGGUGGUGCAUCCCAAUCCCCACACCACUGUACACCUCACUUGUCCACAGAACCCACCAGCUGCGCCCAGCGUAGCGAGACCCAUU .....((.(((......................(((((((((..............)))))))))............................(((((((...)))))))))).)).... (-26.06 = -26.54 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:39 2011