| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,836,624 – 2,836,754 |
| Length | 130 |
| Max. P | 0.998708 |

| Location | 2,836,624 – 2,836,754 |
|---|---|
| Length | 130 |
| Sequences | 13 |
| Columns | 149 |
| Reading direction | forward |
| Mean pairwise identity | 69.46 |
| Shannon entropy | 0.63722 |
| G+C content | 0.49864 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -29.82 |
| Energy contribution | -28.94 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.998708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
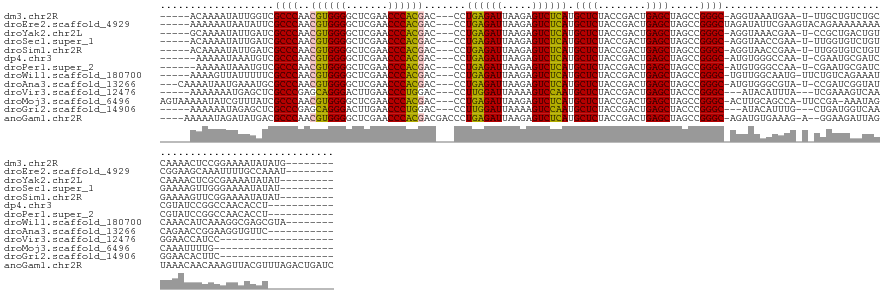
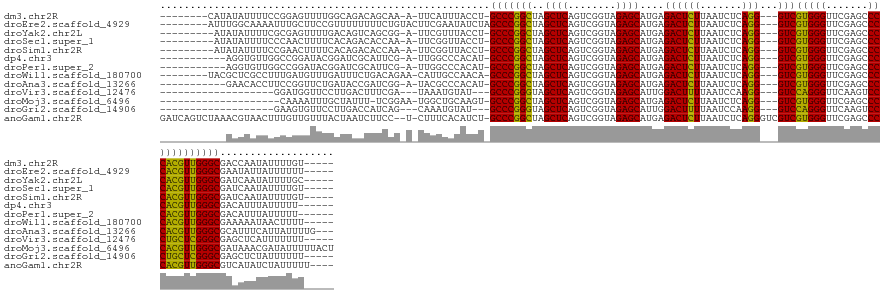
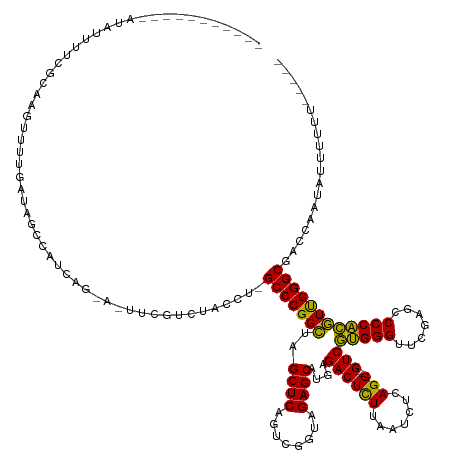
>dm3.chr2R 2836624 130 + 21146708 -----ACAAAAUAUUGGUCGCCCAACGUGGGGCUCGAACCCACGAC---CCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC-AGGUAAAUGAA-U-UUGCUGUCUGCCAAAACUCCGGAAAAUAUAUG-------- -----........(((((.((((..((((((.......))))))..---..((((((.....)))))).((((........)))).....))))-.((((((....-)-)))))....)))))..................-------- ( -38.70, z-score = -2.00, R) >droEre2.scaffold_4929 20279666 133 + 26641161 -----AAAAAAUAAUAUUCGCCCAACGUGGGGCUCGAACCCACGAC---CCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUAGAUAUUCGAAGUACAGAAAAAAAACGGAAGCAAAUUUUGCCAAAU-------- -----...........(((((((..((((((.......))))))..---..((((((.....)))))).((((........)))).....))))......(((........)))........)))(((.....))).....-------- ( -33.70, z-score = -1.94, R) >droYak2.chr2L 15568828 129 + 22324452 -----GCAAAAUAUUGAUCGCCCAACGUGGGGCUCGAACCCACGAC---CCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC-AGGUAAACGAA-U-CCGCUGACUGUCAAAACUCGCGAAAAUAUAU--------- -----((......(((((.((((..((((((.......))))))..---..((((((.....)))))).((((........)))).....))))-.(((...((..-.-.))...)))))))).....))..........--------- ( -36.30, z-score = -1.79, R) >droSec1.super_1 518466 129 + 14215200 -----ACAAAAUAUUGAUCGCCCAACGUGGGGCUCGAACCCACGAC---CCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC-AGGUAACCGAA-U-UUGGUGUCUGUGAAAAGUUGGGAAAAUAUAU--------- -----.....(((((.....(((((((((((.......)))))(((---((((((((.....)))))).((((........))))....(((..-......)))..-.-..)).)))........))))))..)))))..--------- ( -41.30, z-score = -2.35, R) >droSim1.chr2R 1653159 129 + 19596830 -----ACAAAAUAUUGAUCGCCCAACGUGGGGCUCGAACCCACGAC---CCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC-AGGUAACCGAA-U-UUGGUGUCUGUGAAAAGUUCGGAAAAUAUAU--------- -----.....(((((.(((((((..((((((.......))))))..---..((((((.....)))))).((((........)))).....))))-.)))..(((((-(-((..(......)..))))))))..)))))..--------- ( -40.70, z-score = -2.36, R) >dp4.chr3 1125355 126 + 19779522 ------AAAAAUAAAUGUCGCCCAACGUGGGGCUCGAACCCACGAC---CCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC-AUGUGGGCCAA-U-CGAAUGCGAUCCGUAUCCGGCCAACACCU----------- ------.............((((..((((((.......))))))..---..((((((.....)))))).((((........)))).....))))-.(((.((((..-.-.(.(((((...))))).))))).)))...----------- ( -41.60, z-score = -1.97, R) >droPer1.super_2 1289602 126 + 9036312 ------AAAAAUAAAUGUCGCCCAACGUGGGGCUCGAACCCACGAC---CCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC-AUGUGGGCCAA-U-CGAAUGCGAUCCGUAUCCGGCCAACACCU----------- ------.............((((..((((((.......))))))..---..((((((.....)))))).((((........)))).....))))-.(((.((((..-.-.(.(((((...))))).))))).)))...----------- ( -41.60, z-score = -1.97, R) >droWil1.scaffold_180700 5905849 131 + 6630534 -----AAAAGUUAUUUUUCGCCCAACGUGGGGCUCGAACCCACGAC---CCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC-UGUUGGCAAUG-UUCUGUCAGAAAUCAAACAUCAAAGGCGAGCGUA-------- -----...........((((((...((((((.......))))))..---.....((((.(((((.....)))))...(((.((((..(((((..-..)))))...)-))).)))...))))..........))))))....-------- ( -39.70, z-score = -1.15, R) >droAna3.scaffold_13266 9159029 129 + 19884421 ---CAAAAUAAUGAAAUGCGCCCAACGUGGGGCUCGAACCCACGAC---CCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC-AUGUGGGCGUA-U-CCGAUCGGUAUCAGAACCGGAAGGUGUUC----------- ---............(((((((((.((((((.......)))))).(---((((((((.....)))))).((((........)))).....))).-...))))))))-)-((..(((((......)))))..)).....----------- ( -49.30, z-score = -3.06, R) >droVir3.scaffold_12476 8466 116 - 9040 -----AAAAAAAUGAGCUCGCCCGAGCAGGGACUUGAACCCUGGAC---CCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGC---AUACAUUUA---UCGAAAGUCAAGGAACCAUCC------------------- -----..........((((....)))).((..(((((..(((((..---..((((((.....)))))).((((........))))...))))).---.........---.(....))))))...))....------------------- ( -33.40, z-score = -2.33, R) >droMoj3.scaffold_6496 25290770 123 + 26866924 AGUAAAAAUAUCGUUUAUCGCCCAACGUGGGGCUCGAACCCACGAC---CCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC-ACUUGCAGCCA-UUCCGA-AAAUAGCAAAUUUUG-------------------- ....(((((..........((((..((((((.......))))))..---..((((((.....)))))).((((........)))).....))))-..((((.....-......-.....))))))))).-------------------- ( -32.99, z-score = -1.50, R) >droGri2.scaffold_14906 2746829 116 - 14172833 -----AAAAAAUAGAGCUCGCCCGAGCAGGGACUUGAACCCUGGAC---CCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGC---AUACAUUUG---CUGAUGGUCAAGGAACACUUC------------------- -----..........((((....)))).(...(((((.((((((..---..((((((.....)))))).((((........))))...))))((---(......))---)....)))))))...).....------------------- ( -32.10, z-score = -1.18, R) >anoGam1.chr2R 50985799 141 + 62725911 ----AAAAAUAGAUAUGACGCCCAACGUGGGGCUCGAACCCACGACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC-AGAUGUGAAAG-A--GGAAGAUUAGUAAACAACAAAGUUACGUUUAGACUGAUC ----...............((((..((((((.......)))))).......((((((.....)))))).((((........)))).....))))-...........-.--....(((((((....(((........)))...))))))) ( -37.80, z-score = -2.05, R) >consensus _____AAAAAAUAUUGAUCGCCCAACGUGGGGCUCGAACCCACGAC___CCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC_AGGUAAACGAA_U_CUGAUGACUAUCAAAACUUGCGAAAAUAU___________ ...................((((..((((((.......)))))).......((((((.....)))))).((((........)))).....))))....................................................... (-29.82 = -28.94 + -0.89)
| Location | 2,836,624 – 2,836,754 |
|---|---|
| Length | 130 |
| Sequences | 13 |
| Columns | 149 |
| Reading direction | reverse |
| Mean pairwise identity | 69.46 |
| Shannon entropy | 0.63722 |
| G+C content | 0.49864 |
| Mean single sequence MFE | -40.39 |
| Consensus MFE | -31.50 |
| Energy contribution | -30.07 |
| Covariance contribution | -1.43 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.994726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
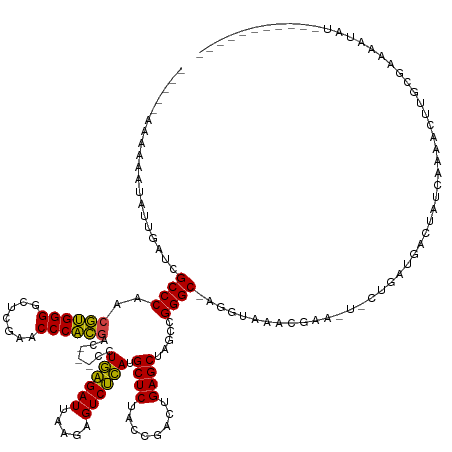
>dm3.chr2R 2836624 130 - 21146708 --------CAUAUAUUUUCCGGAGUUUUGGCAGACAGCAA-A-UUCAUUUACCU-GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGG---GUCGUGGGUUCGAGCCCCACGUUGGGCGACCAAUAUUUUGU----- --------...........((((((.(((((.((......-.-.))........-(((((((..((((........))))....((((((.......)))---)))(((((.......))))))))))))).)))).)))))).----- ( -39.50, z-score = -1.19, R) >droEre2.scaffold_4929 20279666 133 - 26641161 --------AUUUGGCAAAAUUUGCUUCCGUUUUUUUUCUGUACUUCGAAUAUCUAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGG---GUCGUGGGUUCGAGCCCCACGUUGGGCGAAUAUUAUUUUUU----- --------....((((.....))))......................(((((...(((((((..((((........))))....((((((.......)))---)))(((((.......))))))))))))..))))).......----- ( -36.20, z-score = -0.83, R) >droYak2.chr2L 15568828 129 - 22324452 ---------AUAUAUUUUCGCGAGUUUUGACAGUCAGCGG-A-UUCGUUUACCU-GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGG---GUCGUGGGUUCGAGCCCCACGUUGGGCGAUCAAUAUUUUGC----- ---------..........(((((..(((((.((.((((.-.-..)))).))..-(((((((..((((........))))....((((((.......)))---)))(((((.......))))))))))))).))))...)))))----- ( -41.20, z-score = -1.31, R) >droSec1.super_1 518466 129 - 14215200 ---------AUAUAUUUUCCCAACUUUUCACAGACACCAA-A-UUCGGUUACCU-GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGG---GUCGUGGGUUCGAGCCCCACGUUGGGCGAUCAAUAUUUUGU----- ---------.........((((((........(((.((..-.-....(((..((-(((.((((.....)))))))))))).(((((.......)))))))---)))(((((.......)))))))))))...............----- ( -37.40, z-score = -1.53, R) >droSim1.chr2R 1653159 129 - 19596830 ---------AUAUAUUUUCCGAACUUUUCACAGACACCAA-A-UUCGGUUACCU-GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGG---GUCGUGGGUUCGAGCCCCACGUUGGGCGAUCAAUAUUUUGU----- ---------..........((((((...(((.(((.((..-.-....(((..((-(((.((((.....)))))))))))).(((((.......)))))))---)))))))))))).((((......))))..............----- ( -36.20, z-score = -0.95, R) >dp4.chr3 1125355 126 - 19779522 -----------AGGUGUUGGCCGGAUACGGAUCGCAUUCG-A-UUGGCCCACAU-GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGG---GUCGUGGGUUCGAGCCCCACGUUGGGCGACAUUUAUUUUU------ -----------..((((.(((((.((.(((((...)))))-)-)))))).))))-(((((((..((((........))))....((((((.......)))---)))(((((.......)))))))))))).............------ ( -46.60, z-score = -1.45, R) >droPer1.super_2 1289602 126 - 9036312 -----------AGGUGUUGGCCGGAUACGGAUCGCAUUCG-A-UUGGCCCACAU-GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGG---GUCGUGGGUUCGAGCCCCACGUUGGGCGACAUUUAUUUUU------ -----------..((((.(((((.((.(((((...)))))-)-)))))).))))-(((((((..((((........))))....((((((.......)))---)))(((((.......)))))))))))).............------ ( -46.60, z-score = -1.45, R) >droWil1.scaffold_180700 5905849 131 - 6630534 --------UACGCUCGCCUUUGAUGUUUGAUUUCUGACAGAA-CAUUGCCAACA-GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGG---GUCGUGGGUUCGAGCCCCACGUUGGGCGAAAAAUAACUUUU----- --------.....(((((...(((((((...........)))-))))..((((.-((((.....((((........)))).(((((.......)))))))---)).(((((.......))))))))))))))............----- ( -40.00, z-score = -0.88, R) >droAna3.scaffold_13266 9159029 129 - 19884421 -----------GAACACCUUCCGGUUCUGAUACCGAUCGG-A-UACGCCCACAU-GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGG---GUCGUGGGUUCGAGCCCCACGUUGGGCGCAUUUCAUUAUUUUG--- -----------(((.....(((((((........))))))-)-..(((((((((-((((((((.....))))))....))))).((((((.......)))---)))(((((.......)))))..))))))...))).........--- ( -45.90, z-score = -2.23, R) >droVir3.scaffold_12476 8466 116 + 9040 -------------------GGAUGGUUCCUUGACUUUCGA---UAAAUGUAU---GCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGG---GUCCAGGGUUCAAGUCCCUGCUCGGGCGAGCUCAUUUUUUU----- -------------------(((((((((.(((.....)))---.........---(((((((((((((........))))..(((((((((.....))))---)))))(((.......)))))))))))))))).)))))....----- ( -40.70, z-score = -2.07, R) >droMoj3.scaffold_6496 25290770 123 - 26866924 --------------------CAAAAUUUGCUAUUU-UCGGAA-UGGCUGCAAGU-GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGG---GUCGUGGGUUCGAGCCCCACGUUGGGCGAUAAACGAUAUUUUUACU --------------------........(((((((-...)))-))))......(-(((((((..((((........))))....((((((.......)))---)))(((((.......))))))))))))).................. ( -37.70, z-score = -0.90, R) >droGri2.scaffold_14906 2746829 116 + 14172833 -------------------GAAGUGUUCCUUGACCAUCAG---CAAAUGUAU---GCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGG---GUCCAGGGUUCAAGUCCCUGCUCGGGCGAGCUCUAUUUUUU----- -------------------..((.((((.(((.....)))---.........---(((((((((((((........))))..(((((((((.....))))---)))))(((.......)))))))))))))))).)).......----- ( -39.40, z-score = -1.95, R) >anoGam1.chr2R 50985799 141 - 62725911 GAUCAGUCUAAACGUAACUUUGUUGUUUACUAAUCUUCC--U-CUUUCACAUCU-GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUCGUGGGUUCGAGCCCCACGUUGGGCGUCAUAUCUAUUUUU---- (((.(((...((((......))))....))).)))....--.-...........-(((((((..((((........))))..((((((((.......)))))).))(((((.......))))))))))))...............---- ( -37.70, z-score = -1.30, R) >consensus ___________AUAUUUUCGCAAGUUUUGAUAGCCAUCAG_A_UUCGUCUACCU_GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGG___GUCGUGGGUUCGAGCCCCACGUUGGGCGACCAAUAUUUUUU_____ .......................................................(((((((..((((........)))).(((((.......)))))........(((((.......))))))))))))................... (-31.50 = -30.07 + -1.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:36 2011