
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,835,782 – 2,835,865 |
| Length | 83 |
| Max. P | 0.994931 |

| Location | 2,835,782 – 2,835,865 |
|---|---|
| Length | 83 |
| Sequences | 12 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Shannon entropy | 0.40177 |
| G+C content | 0.56203 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -26.37 |
| Energy contribution | -25.82 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.994931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
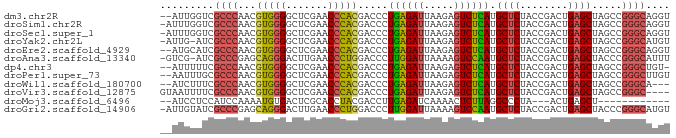
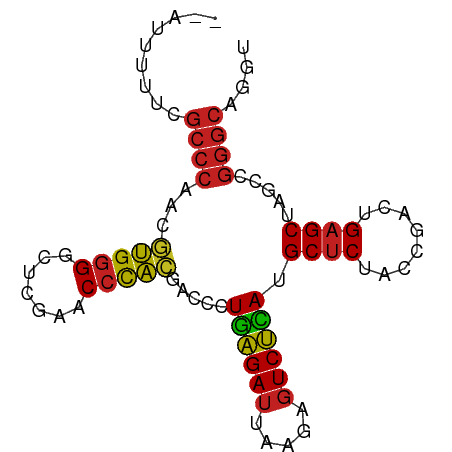
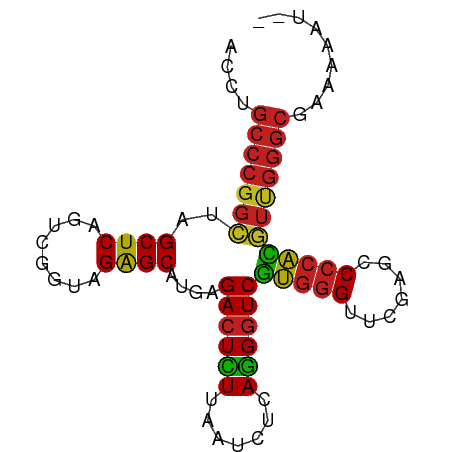
>dm3.chr2R 2835782 83 + 21146708 --AUUGGUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGU --.......((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).... ( -31.50, z-score = -1.71, R) >droSim1.chr2R 1652386 84 + 19596830 -AUUUGGUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGU -........((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).... ( -31.50, z-score = -1.71, R) >droSec1.super_1 517707 84 + 14215200 -AUUUGGUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGU -........((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).... ( -31.50, z-score = -1.71, R) >droYak2.chr2L 15567975 83 + 22324452 -AUUG-AUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGU -....-...((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).... ( -31.50, z-score = -2.91, R) >droEre2.scaffold_4929 6926336 83 - 26641161 --AUGCAUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGU --.......((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).... ( -31.50, z-score = -2.10, R) >droAna3.scaffold_13340 18293481 83 - 23697760 -GUCG-AUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUUU -....-...(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).... ( -29.10, z-score = -2.83, R) >dp4.chr3 1802541 82 - 19779522 --AUUUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGU- --.......((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))...- ( -31.00, z-score = -2.95, R) >droPer1.super_73 78250 83 + 267513 --AAUUUGCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUUGU --.......((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).... ( -31.00, z-score = -2.81, R) >droWil1.scaffold_180700 5897142 80 + 6630534 --AUCUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCA--- --.......((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).--- ( -31.50, z-score = -3.25, R) >droVir3.scaffold_12875 15830063 81 - 20611582 GUAAUUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC---- .........((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))---- ( -29.80, z-score = -2.86, R) >droMoj3.scaffold_6496 25958194 68 - 26866924 --AUCCUCCAUCCAAAAUGUGACUCGCACCUACGACCUUGAGAUCAAAACUCUUAGGCCCUA---ACUGAGCU------------ --...(((..........(((.....))).......(((((((.......))))))).....---...)))..------------ ( -6.80, z-score = 0.26, R) >droGri2.scaffold_14906 2742463 84 - 14172833 -AUUGUAUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUGU -........(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).... ( -29.10, z-score = -2.83, R) >consensus __AUUUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGU .........((((...(((((.......))))).....((((((.....)))))).((((........)))).....)))).... (-26.37 = -25.82 + -0.55)
| Location | 2,835,782 – 2,835,865 |
|---|---|
| Length | 83 |
| Sequences | 12 |
| Columns | 85 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Shannon entropy | 0.40177 |
| G+C content | 0.56203 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -29.51 |
| Energy contribution | -29.01 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
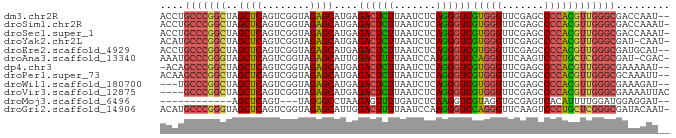
>dm3.chr2R 2835782 83 - 21146708 ACCUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGACCAAU-- ...((((((((..((((........))))....((((((.......))))))(((((.......)))))))))))))......-- ( -32.80, z-score = -1.64, R) >droSim1.chr2R 1652386 84 - 19596830 ACCUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGACCAAAU- ...((((((((..((((........))))....((((((.......))))))(((((.......))))))))))))).......- ( -32.80, z-score = -1.72, R) >droSec1.super_1 517707 84 - 14215200 ACCUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGACCAAAU- ...((((((((..((((........))))....((((((.......))))))(((((.......))))))))))))).......- ( -32.80, z-score = -1.72, R) >droYak2.chr2L 15567975 83 - 22324452 ACAUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAU-CAAU- ...((((((((..((((........))))....((((((.......))))))(((((.......)))))))))))))..-....- ( -33.00, z-score = -2.24, R) >droEre2.scaffold_4929 6926336 83 + 26641161 ACCUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAUGCAU-- ...((((((((..((((........))))....((((((.......))))))(((((.......)))))))))))))......-- ( -32.80, z-score = -1.41, R) >droAna3.scaffold_13340 18293481 83 + 23697760 AAAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAU-CGAC- ...((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))..-....- ( -35.50, z-score = -3.49, R) >dp4.chr3 1802541 82 + 19779522 -ACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAAAU-- -...(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))).......-- ( -32.90, z-score = -2.58, R) >droPer1.super_73 78250 83 - 267513 ACAAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGCAAAUU-- ....(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))).......-- ( -32.90, z-score = -2.48, R) >droWil1.scaffold_180700 5897142 80 - 6630534 ---UGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAGAU-- ---.(((((((..((((........))))....((((((.......))))))(((((.......)))))))))))).......-- ( -32.70, z-score = -2.19, R) >droVir3.scaffold_12875 15830063 81 + 20611582 ----GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAAUUAC ----(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))......... ( -32.70, z-score = -2.64, R) >droMoj3.scaffold_6496 25958194 68 + 26866924 ------------AGCUCAGU---UAGGGCCUAAGAGUUUUGAUCUCAAGGUCGUAGGUGCGAGUCACAUUUUGGAUGGAGGAU-- ------------..(((..(---((.(((((..(((.......))).))))).)))((.((((......)))).)).)))...-- ( -14.80, z-score = 0.20, R) >droGri2.scaffold_14906 2742463 84 + 14172833 ACAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAUACAAU- ...((((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))).......- ( -35.50, z-score = -3.79, R) >consensus ACCUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAAAU__ ....(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))......... (-29.51 = -29.01 + -0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:34 2011