
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,834,282 – 2,834,379 |
| Length | 97 |
| Max. P | 0.998668 |

| Location | 2,834,282 – 2,834,379 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 69.11 |
| Shannon entropy | 0.69083 |
| G+C content | 0.45602 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -22.97 |
| Energy contribution | -21.45 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.44 |
| SVM RNA-class probability | 0.998668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
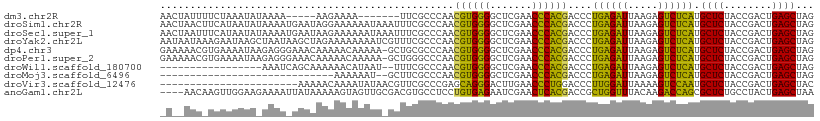
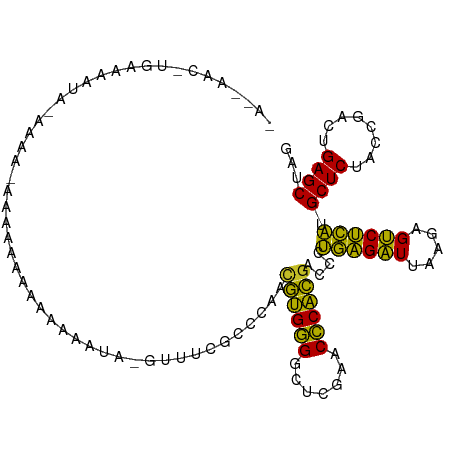
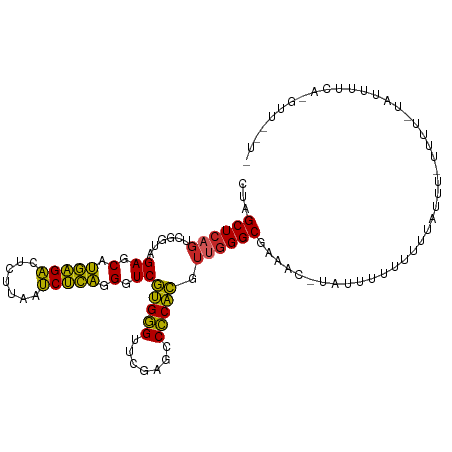
>dm3.chr2R 2834282 97 + 21146708 AACUAUUUUCUAAAUAUAAAA-----AAGAAAA-------UUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAG ....(((((((..........-----.))))))-------)........((((((.......))))))....((((((.....)))))).((((........))))... ( -24.50, z-score = -3.18, R) >droSim1.chr2R 1651034 109 + 19596830 AACUAACUUCAUAAUAUAAAAUGAAUAGGAAAAAAUAAAUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAG ..(((..(((((........))))))))((((.......))))......((((((.......))))))....((((((.....)))))).((((........))))... ( -25.60, z-score = -2.83, R) >droSec1.super_1 516330 109 + 14215200 AACUAAUUUCAUAAUAUAAAAUGAAUAAGAAAAAAUAAAUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAG .......(((((........)))))...((((.......))))......((((((.......))))))....((((((.....)))))).((((........))))... ( -24.70, z-score = -3.09, R) >droYak2.chr2L 15566878 109 + 22324452 AAUAAUAAAGAAUAAGCUAAUAAGCUAGAAAAAAAAUCGUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAG ..............((((....)))).((((........))))......((((((.......))))))....((((((.....)))))).((((........))))... ( -26.30, z-score = -2.37, R) >dp4.chr3 1124437 108 + 19779522 GAAAAACGUGAAAAUAAGAGGGAAACAAAAACAAAAA-GCUGCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAG ..................((((...............-((...))....((((((.......)))))).))))(((((.....)))))..((((........))))... ( -26.90, z-score = -2.08, R) >droPer1.super_2 1288685 108 + 9036312 GAAAAACGUGAAAAUAAGAGGGAAACAAAAACAAAAA-GCUGGGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAG ......((((..........(....)...........-..(((((((......)))))))....))))....((((((.....)))))).((((........))))... ( -29.10, z-score = -2.28, R) >droWil1.scaffold_180700 5874055 90 + 6630534 -----------------AAAUCAGCAAAAAACAUAAU--UUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAG -----------------......((.((((......)--))).))....((((((.......))))))....((((((.....)))))).((((........))))... ( -23.40, z-score = -2.66, R) >droMoj3.scaffold_6496 25957777 78 - 26866924 -----------------------------AAAAAAU--GCUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAG -----------------------------.......--...........((((((.......))))))....((((((.....)))))).((((........))))... ( -22.50, z-score = -2.51, R) >droVir3.scaffold_12476 2639 86 - 9040 -----------------------AAAAACAAAAUAUAACGUUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAC -----------------------................(((((..((..(((((.......)))))((.(.((((((.....)))))).).))...))..)))))... ( -20.10, z-score = -2.14, R) >anoGam1.chr2L 44194329 105 + 48795086 ----AACAAGUUGGAAGAAAAUUAUAAAAAGUAGUUGCGACGUGCCUCCUGUGAGAAUCGAACUCACGACCGCUGGUUUACAAGACCAGCGCUCUGCCUACUGAGCUAA ----.....((((((....((((((.....))))))((.....)).))).(((((.......)))))))).(((((((.....)))))))((((........))))... ( -26.70, z-score = -1.41, R) >consensus _A__AAC_UGAAAAUA_AAAA_AAAAAAAAAAAAAUA_GUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAG .................................................((((((.......))))))....((((((.....)))))).((((........))))... (-22.97 = -21.45 + -1.52)
| Location | 2,834,282 – 2,834,379 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 69.11 |
| Shannon entropy | 0.69083 |
| G+C content | 0.45602 |
| Mean single sequence MFE | -26.36 |
| Consensus MFE | -22.75 |
| Energy contribution | -21.61 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.37 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
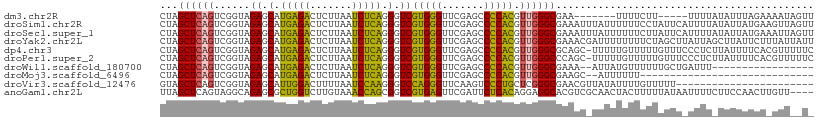
>dm3.chr2R 2834282 97 - 21146708 CUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAA-------UUUUCUU-----UUUUAUAUUUAGAAAAUAGUU ...((((((.(....((.(.(((((.......))))).).))(((((.......))))))))))))..(-------((((((.-----..........))))))).... ( -26.10, z-score = -1.24, R) >droSim1.chr2R 1651034 109 - 19596830 CUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAUUUAUUUUUUCCUAUUCAUUUUAUAUUAUGAAGUUAGUU ((((((((....(((((..((((((((((.......))))))(((((.......)))))..((((.((((.....)))).)))).)))))))))....)).)))))).. ( -28.90, z-score = -1.59, R) >droSec1.super_1 516330 109 - 14215200 CUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAUUUAUUUUUUCUUAUUCAUUUUAUAUUAUGAAAUUAGUU ...((((((.(....((.(.(((((.......))))).).))(((((.......))))))))))))((((.......))))...(((((........)))))....... ( -26.30, z-score = -1.11, R) >droYak2.chr2L 15566878 109 - 22324452 CUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAACGAUUUUUUUUCUAGCUUAUUAGCUUAUUCUUUAUUAUU ...((((((.(....((.(.(((((.......))))).).))(((((.......))))))))))))((((........)))).((((....)))).............. ( -26.00, z-score = -0.08, R) >dp4.chr3 1124437 108 - 19779522 CUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGCAGC-UUUUUGUUUUUGUUUCCCUCUUAUUUUCACGUUUUUC ...((((........)))).(((((((((.......))))))(.(((..(((((((......))))((((.-...))))..)))...))).)......)))........ ( -25.90, z-score = -0.22, R) >droPer1.super_2 1288685 108 - 9036312 CUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCCCAGC-UUUUUGUUUUUGUUUCCCUCUUAUUUUCACGUUUUUC ...((((........)))).(((((.......)))))(((..(((((.......)))))((((....))))-...............)))................... ( -25.60, z-score = -0.23, R) >droWil1.scaffold_180700 5874055 90 - 6630534 CUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAA--AUUAUGUUUUUUGCUGAUUU----------------- .......((((((((((((((((((((((.......))))))(((((.......)))))...........--.))))))..)))))))))).----------------- ( -27.80, z-score = -1.45, R) >droMoj3.scaffold_6496 25957777 78 + 26866924 CUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAGC--AUUUUUU----------------------------- ...(((..(((....((.(.(((((.......))))).).))(((((.......)))))....)))..)))--.......----------------------------- ( -23.30, z-score = -0.67, R) >droVir3.scaffold_12476 2639 86 + 9040 GUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAACGUUAUAUUUUGUUUUU----------------------- (((((....(((.(.(((((.(((((((((.....)))))))))(((.......)))))))).).)))..)))))...........----------------------- ( -26.60, z-score = -2.41, R) >anoGam1.chr2L 44194329 105 - 48795086 UUAGCUCAGUAGGCAGAGCGCUGGUCUUGUAAACCAGCGGUCGUGAGUUCGAUUCUCACAGGAGGCACGUCGCAACUACUUUUUAUAAUUUUCUUCCAACUUGUU---- .......(((((((.((.(((((((.......))))))).))(((((.......)))))............))..))))).........................---- ( -27.10, z-score = -0.91, R) >consensus CUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAC_UAUUUUUUUUUAUUU_UUUU_UAUUUUCA_GUU__U_ ...((((((......((.(.(((((.......))))).).))(((((.......))))).))))))........................................... (-22.75 = -21.61 + -1.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:31 2011