| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,829,324 – 2,829,436 |
| Length | 112 |
| Max. P | 0.993913 |

| Location | 2,829,324 – 2,829,436 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 78.72 |
| Shannon entropy | 0.38741 |
| G+C content | 0.48784 |
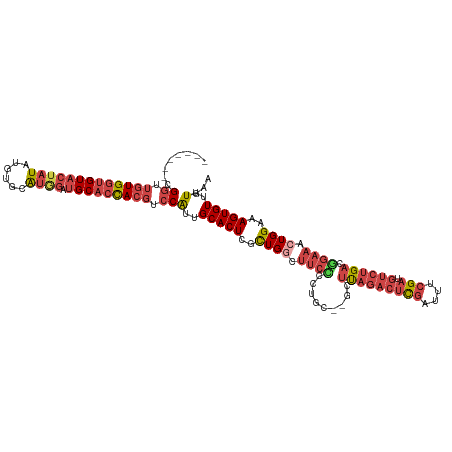
| Mean single sequence MFE | -37.92 |
| Consensus MFE | -25.58 |
| Energy contribution | -28.03 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.993913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
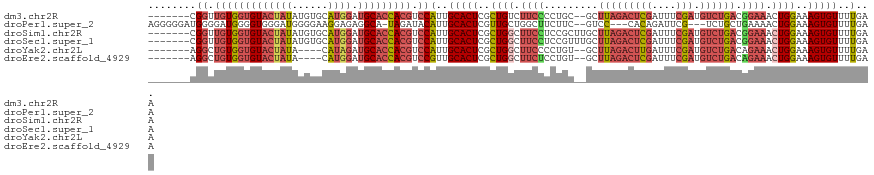
>dm3.chr2R 2829324 112 - 21146708 -------CGGUUGUGGUGUACUAUAUGUGCAUGGAUGCACCACGUCCAUUGCACUCGCUGUCUUCCCCUGC--GCUUAGACUCGAUUUCGAUGUCUGACGGAAACUGGAAAGUGUUUUGAA -------.((.(((((((((((((......)))).))))))))).))...(((((..(((..((((.....--(.(((((((((....))).)))))))))))..)))..)))))...... ( -36.00, z-score = -2.13, R) >droPer1.super_2 1284656 112 - 9036312 AGGGGGAUGGGGAUGGGGUGGGAUGGGGAAGGAGAGGCA-UAGAUACAUUGCACUCGUUGCUGGCUUCUUC--GUCC---CACAGAUUCG---UCUGCUGAAAACUGGAAAGUGUUUUGAA .((.(((((((......(((((((((((((((.((((((-.........))).)))....)...)))))))--))))---)))...))))---))).))(((((((....))).))))... ( -38.20, z-score = -2.07, R) >droSim1.chr2R 1645664 114 - 19596830 -------CGGUUGUGGUGUACUAUAUGUGCAUGGAUGCACCACGUCCAUUGCACUCGCUGGCUUCCUCCGCUUGCUUAGACUCGAUUUCGAUGUCUGACGGAAACUGGAAAGUGUUUUGAA -------.((.(((((((((((((......)))).))))))))).))...(((((..(..(.((((...(....)(((((((((....))).)))))).)))).)..)..)))))...... ( -40.50, z-score = -3.19, R) >droSec1.super_1 511229 114 - 14215200 -------CGGUUGUGGUGUACUAUAUGUGCAUGGAUGCACCACGUCCAUUGCACUCGCUGGCUUCCUCCGUUUGCUUAGACUCGAUUUCGAUGUCUGACGGAAACUGGAAAGUGUUUUGAA -------.((.(((((((((((((......)))).))))))))).))...(((((..(..(.((((...(....)(((((((((....))).)))))).)))).)..)..)))))...... ( -40.30, z-score = -3.21, R) >droYak2.chr2L 15561835 108 - 22324452 -------AGGCUGUGGUGUACUAUA----CAUAGAUGCACCACGUCCAUUGCACUCGCUGGCUUCCCCUGU--GCUUAGACUUGAUUUCGAUGUCUGACAGAAACUGGAAAGUGUUUUGAA -------.(((.(((((((((((..----..))).)))))))))))((..(((((..(..(.(((.....(--(.(((((((((....))).))))))))))).)..)..)))))..)).. ( -35.70, z-score = -2.94, R) >droEre2.scaffold_4929 6920507 108 + 26641161 -------AGGCUGUGGUGUACUAUA----CAUGGAUGCACCACGUCCGUUGCACUCGCUGGCUUCUCCUGU--GCUUAGACUCGAUUUCGAUGUCUGACAGAAACUGGAAAGUGUUUUGAA -------.(((.(((((((((((..----..))).)))))))))))((..(((((..(..(.((((.....--(.(((((((((....))).))))))))))).)..)..)))))..)).. ( -36.80, z-score = -2.62, R) >consensus _______CGGUUGUGGUGUACUAUAUGUGCAUGGAUGCACCACGUCCAUUGCACUCGCUGGCUUCCCCUGC__GCUUAGACUCGAUUUCGAUGUCUGACGGAAACUGGAAAGUGUUUUGAA ........((.(((((((((((((......)))).))))))))).))(..(((((..((((.((((.........(((((((((....))).)))))).)))).))))..)))))..)... (-25.58 = -28.03 + 2.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:30 2011