| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,823,945 – 2,824,074 |
| Length | 129 |
| Max. P | 0.984538 |

| Location | 2,823,945 – 2,824,074 |
|---|---|
| Length | 129 |
| Sequences | 6 |
| Columns | 141 |
| Reading direction | forward |
| Mean pairwise identity | 71.23 |
| Shannon entropy | 0.53249 |
| G+C content | 0.42559 |
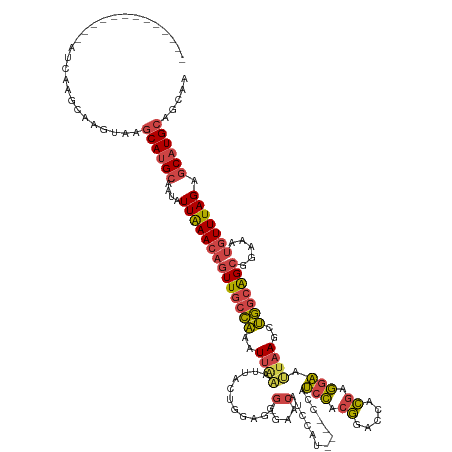
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -21.08 |
| Energy contribution | -21.42 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
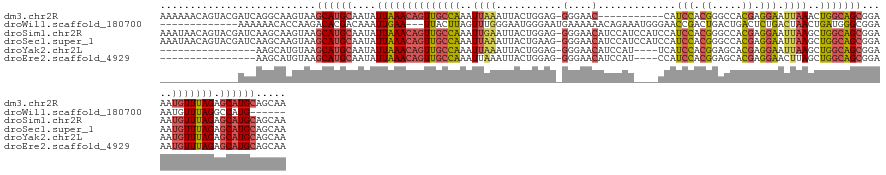
>dm3.chr2R 2823945 129 + 21146708 AAAAAACAGUACGAUCAGGCAAGUAAGCAUGCAAUAUUAAACAGUUGCCAAAUUAAAUUACUGGAG-GGGAAC-----------CAUCCACGGGCCACGAGGAAUUAAACUGGCAGCGGAAAUGUUUAGAGCAUGCAGCAA ..........................((((((....((((((((((((((..((((.........(-(....)-----------).(((.((.....)).))).))))..))))))).....))))))).))))))..... ( -32.60, z-score = -1.98, R) >droWil1.scaffold_180700 2344006 119 + 6630534 -------------AAAAAACACCAAGACACGACAAAUUGAA---UUACUUAGUUUGGGAAUGGGAAUGAAAAAACAGAAAUGGGAACCGACUGACUGACUCUGACUAACUGAUGGGCGGAAAUGUUUAGGCCAUG------ -------------...................((((((((.---....))))))))...((((...........((....))....((....(((....((((.(((.....))).))))...)))..)))))).------ ( -16.60, z-score = 0.61, R) >droSim1.chr2R 1640240 140 + 19596830 AAAUAACAGUACGAUCAAGCAAGUAAGCAUGCAAUAUUAAACAGUUGCCAAAUUGAAUUACUGGAG-GGGAACAUCCAUCCAUCCAUCCACGGGCCACGAGGAAUUAAGCUGGCAGCGGAAAUGUUUAGAGCAUGCAGCAA ..........................((((((....((((((((((((((..((((.....((((.-.(((.......))).))))(((.((.....)).))).))))..))))))).....))))))).))))))..... ( -37.30, z-score = -1.89, R) >droSec1.super_1 505771 140 + 14215200 AAAUAACAGUACGAUCAAGCAAGUAAGCAUGCAAUAUUAAACAGUUGCCAAAUUAAAUUACUGAAG-GGGAACAUCCAUCCAUCCAUCCACGGGCCACGAGGAAUUAAGCUGGCAGCGGAAAUGUUUAGAGCAUGCAGCAA ..........................((((((....((((((((((((((..((((.........(-((((.......)))..)).(((.((.....)).))).))))..))))))).....))))))).))))))..... ( -32.80, z-score = -1.16, R) >droYak2.chr2L 15556946 120 + 22324452 ----------------AAGCAUGUAAGCAUGCAAUAUUAAACAGUUGCCAAAUUAAAUUACUGGAG-GGGAACAUCCAU----UCAUCCACGGAGCACGAGGAAUUAAGCUGGCAGCGGAAAUGUUUAGAGCAUGCAGCAA ----------------.....(((..((((((....((((((((((((((..((((.....((((.-(....).)))).----...(((.((.....)).))).))))..))))))).....))))))).)))))).))). ( -35.60, z-score = -3.03, R) >droEre2.scaffold_4929 6915246 120 - 26641161 ----------------AAGCAUGUAAGCAUGCAAUAUUAAACAGUUGCCAAAUUAAAUUACUGGAG-GGGAACAUCCAU----CCAUCCACGGAGCACGAGGAACUUAGCUGGCAGCGGAAAUGUUUAGAGCAUGCAGCAA ----------------.....(((..((((((....((((((((((((((...........((((.-.((.....)).)----)))(((.((.....)).))).......))))))).....))))))).)))))).))). ( -35.10, z-score = -2.35, R) >consensus _____________AUCAAGCAAGUAAGCAUGCAAUAUUAAACAGUUGCCAAAUUAAAUUACUGGAG_GGGAACAUCCAU____CCAUCCACGGACCACGAGGAAUUAAGCUGGCAGCGGAAAUGUUUAGAGCAUGCAGCAA ..........................((((((....((((((((((((((..((((...........(....).............(((.((.....)).))).))))..))))))).....))))))).))))))..... (-21.08 = -21.42 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:29 2011