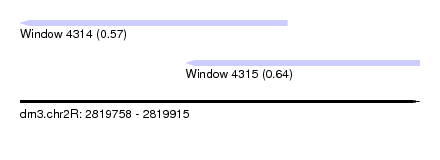
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,819,758 – 2,819,915 |
| Length | 157 |
| Max. P | 0.636980 |

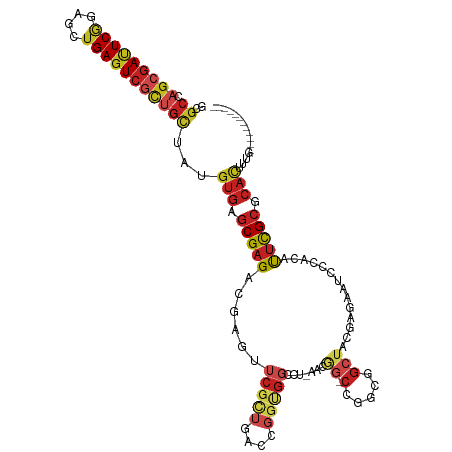
| Location | 2,819,758 – 2,819,863 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.63 |
| Shannon entropy | 0.48538 |
| G+C content | 0.61691 |
| Mean single sequence MFE | -40.95 |
| Consensus MFE | -21.89 |
| Energy contribution | -22.68 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2819758 105 - 21146708 GCGCCAGCGAUUCGGAGCUGAGUCGCUGCUAUGUGAGCGAGACGAGUUCGCUGACCGGUGGCCU-AACGG-CCGGCGGCUACGAGAAUCCCACUUUCGCGCACUUUG------------ ((((((((((((((....))))))))))....(((((((((.....))))))..(((.(((((.-...))-))).)))............)))....))))......------------ ( -47.10, z-score = -2.35, R) >droSim1.chr2R 1636214 107 - 19596830 GCGCCAGCGAUUCAGAGCUGAGUCGCUGCUAUGUGAGCGAGACGAGUUCGCUGACCGGUGGCCUAAACGGCCCGGCGGCUACGAGAAUCCCACUUUCGCGCACUUCG------------ ((((((((((((((....))))))))))....(((.(.((..(..((..((((.((((..(((.....))))))))))).))..)..))))))....))))......------------ ( -44.20, z-score = -1.59, R) >droSec1.super_1 501807 105 - 14215200 GCGCCAGCGAUUCGGAGCUGAGUCGCUGCUAUGUGAGCGAGACGAGUUCGCUGACCGGUGGCCU-AACGG-CCGGCGGCUAUGAGAAUCCCACUUUCGCGCACUUCG------------ ((((((((((((((....))))))))))....(((((((((.....))))))..(((.(((((.-...))-))).)))............)))....))))......------------ ( -47.10, z-score = -2.17, R) >droYak2.chr2L 15552988 105 - 22324452 GCGCCAGCGAUUCGGAGCUGAGUCGCUGCUAUGUGAGCGAGACGAGUUCGCUGACCGGUGGUCU-AACGG-CCGGCGGCUACGAGAAUCCCACUUUCGCGCACUUUG------------ ((((((((((((((....))))))))))....(((((((((.....))))))..(((.(((((.-...))-))).)))............)))....))))......------------ ( -44.40, z-score = -1.57, R) >droEre2.scaffold_4929 6911124 105 + 26641161 GCGCCAGCGAUUCGGAGCUGAGUCGCUGCUAUGUGAGCGAGACGAGUUCGCUAACCGGUGGCCU-GACGG-CCGGCGGCUAUGAGAAUCCCACUUUCGCGCACUUUG------------ ((((((((((((((....))))))))))....(((((((((.....))))))..(((.(((((.-...))-))).)))............)))....))))......------------ ( -45.10, z-score = -1.94, R) >droAna3.scaffold_13266 9145767 105 - 19884421 GAGCCAGCGAUUCGGAGCUGAGUCGUUGCUAUGUGAGCGAGACGAGUUCGCUGACCGGCGGGAU-GACGG-CCGGCGGCUACGAGAAUCCCACAUUUGCGCACUUUG------------ ..((.(((((((((....)))))))))((.(((((.(.((..(..((..((((.(((((.(...-..).)-)))))))).))..)..))))))))..))))......------------ ( -40.10, z-score = -1.09, R) >dp4.chr3 1099452 105 - 19779522 GCGCCAGCGAUUCGGAGCUGAGUCGCUGCUAUGUGAGCGAGACGAGUUCGCUGACCGGCGGCCU-GACAG-CGGGCGGCUACGAGAACCCCACGUUCGCCCACUUUG------------ ..((((((((((((....)))))))))(((..((.((((((.....)))))).)).))))))..-...((-.((((((..(((.(.....).))))))))).))...------------ ( -42.30, z-score = -0.71, R) >droPer1.super_65 158385 85 - 397436 ------GCCA-UCAUGCUGGAGUCUCAGCU-CAUCGACGUGGUCUGCUCGGCGGGCCAUGCCACU------ACUGCGACCUC----AUCCCACACCCGCGCUG---------------- ------.(((-......))).....((((.-....(.((((((((((...)))))))))).)...------...(((.....----..........)))))))---------------- ( -23.26, z-score = 0.93, R) >droWil1.scaffold_180700 2338769 102 - 6630534 AUGCCAGCGAUUCGGAGCUGAGUCGUUGCUAUGUGAGCGAGACGAGUUCGCUGA-----------------CCGCUGGCUAUGAGAAUCCCACAUUUGCCCAUUUAUCCCGCGAAGAUC ..(((((((.((.((((((..(((.(((((.....)))))))).))))).).))-----------------.)))))))...............(((((...........))))).... ( -30.50, z-score = -0.90, R) >droVir3.scaffold_12823 826543 105 + 2474545 AUGCCUCGGACUCGGAGCUGAGUCGCUGUUAUGUGAGCGAGACGAGCUCGCUGACCGGCGGCAU-AUUGG-CCGGCGGCUAUGAGAAUCCGACAUUCGCGCACUUUG------------ .(((((((((((((((((((..(((((........)))))..).)))))((((.(((((.(...-..).)-))))))))..))))..))))).....).))).....------------ ( -39.90, z-score = -0.71, R) >droMoj3.scaffold_6496 25937225 105 + 26866924 GCGCCAGCGACUCGGAGCUGAGUCGCUGCUACGUGAGCGAGACGAGCUCGUUGACGGGCGGACU-CCUGG-CCGGGGGCUACGAGAAUCCCACAUUCGCGCACUUUG------------ ((((.(((((((((....)))))))))(((.(((.((((((.....)))))).))))))(((((-(.(((-((...))))).)))..))).......))))......------------ ( -48.00, z-score = -1.97, R) >droGri2.scaffold_15245 16383594 105 - 18325388 GCGCCAGCGAUUCGGAGCUGAGUCGCUGCUAUGUGAGCGAGACCAGUUCACUGACUGGCGGCAU-ACUGG-CCGGUGGCUAUGAGAAUCCCACAUUUGCGCAUUUUG------------ ((((((((((((((....))))))))))..(((((.((..(.((((((....))))))).))..-..(((-((...))))).........)))))..))))......------------ ( -39.40, z-score = -1.33, R) >consensus GCGCCAGCGAUUCGGAGCUGAGUCGCUGCUAUGUGAGCGAGACGAGUUCGCUGACCGGUGGCCU_AACGG_CCGGCGGCUACGAGAAUCCCACAUUCGCGCACUUUG____________ .(((((((((((((....)))))))))(((..((.((((((.....)))))).)).)))..............)))).......................................... (-21.89 = -22.68 + 0.79)


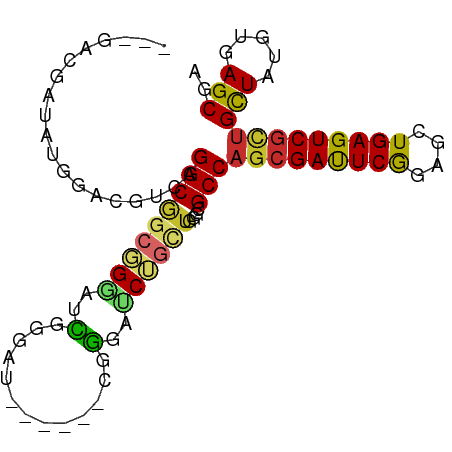
| Location | 2,819,823 – 2,819,915 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 72.03 |
| Shannon entropy | 0.59301 |
| G+C content | 0.63306 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -18.81 |
| Energy contribution | -18.71 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.636980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2819823 92 - 21146708 ---GACGAUAUGGACGUCAGGCGGCGGGAUCGGGAUCGGGACCGGGAUCUGCUGGGCGCCAGCGAUUCGGAGCUGAGUCGCUGCUAUGUGAGCGA ---....((((((.((((.((((((((..(((....)))..)))....))))).)))).((((((((((....))))))))))))))))...... ( -36.20, z-score = -0.51, R) >droSim1.chr2R 1636281 92 - 19596830 ---GACGAUAUGGACGUCAGGCGGCGGGAUCGGGAUCGGGACCGGGAUCUGCUGGGCGCCAGCGAUUCAGAGCUGAGUCGCUGCUAUGUGAGCGA ---....((((((.((((.((((((((..(((....)))..)))....))))).)))).((((((((((....))))))))))))))))...... ( -37.00, z-score = -0.92, R) >droSec1.super_1 501872 86 - 14215200 ---GAAGAUAUGGACGUCAGGCGGCGGGAUCGGGAU------CGGGAUCUGCUGGGCGCCAGCGAUUCGGAGCUGAGUCGCUGCUAUGUGAGCGA ---...........(((((((((((((..((((..(------(.(((((.(((((...)))))))))).)).)))).))))))))...))).)). ( -34.90, z-score = -1.34, R) >droYak2.chr2L 15553053 86 - 22324452 ---GACGAUAUGGACGUCAGGCAGCGGGAUCGAGAU------CGGGAUCUGCUCGGCGCCAGCGAUUCGGAGCUGAGUCGCUGCUAUGUGAGCGA ---((((.......)))).((((((((((((.....------...)))))(((((((.((........)).)))))))))))))).......... ( -34.30, z-score = -0.64, R) >droEre2.scaffold_4929 6911189 86 + 26641161 ---GACGAAAUGGACGUCAGGCGGCGGGAUCGAGAC------CGGGAUCUGCUGGGCGCCAGCGAUUCGGAGCUGAGUCGCUGCUAUGUGAGCGA ---((((.......)))).((((((((..((.((..------(.(((((.(((((...)))))))))).)..)))).)))))))).......... ( -32.90, z-score = -0.46, R) >droAna3.scaffold_13266 9145832 95 - 19884421 GACGAGAGUGCGGAGGCACGGCGACGGGAUCGGGAUAGGGACAGGGACCUGCUUGGAGCCAGCGAUUCGGAGCUGAGUCGUUGCUAUGUGAGCGA .((....))((....(((..(((((((..(((((.((((........)))))))))..))...((((((....)))))))))))..)))..)).. ( -31.70, z-score = -0.41, R) >droVir3.scaffold_12823 826608 86 + 2474545 --GGCAUCGAUGGCCAAUUGGCCGCGGAACGACGG-------CAUCAGCAAUUGAAUGCCUCGGACUCGGAGCUGAGUCGCUGUUAUGUGAGCGA --.((.(((..((((....))))((((..(((.((-------((((((...))).))))))))((((((....)))))).))))....))))).. ( -31.20, z-score = -0.73, R) >droMoj3.scaffold_6496 25937290 74 + 26866924 -------------------GGCGACGGCGAUGCGCAGCGG--CAGCGUCAACUGGGCGCCAGCGACUCGGAGCUGAGUCGCUGCUACGUGAGCGA -------------------((((.(((.(((((((....)--).)))))..)))..))))(((((((((....)))))))))(((.....))).. ( -34.60, z-score = -0.37, R) >droGri2.scaffold_15245 16383659 86 - 18325388 --GAUGAAAGUGGAGGAUUGGCGGCUGCCCGACGU-------CAUCAGCAGUUGGGCGCCAGCGAUUCGGAGCUGAGUCGCUGCUAUGUGAGCGA --.......((.......(((((((.(((((((..-------........))))))))))(((((((((....))))))))))))).....)).. ( -34.70, z-score = -1.30, R) >consensus ___GACGAUAUGGACGUCAGGCGGCGGGAUCGGGAU______CGGGAUCUGCUGGGCGCCAGCGAUUCGGAGCUGAGUCGCUGCUAUGUGAGCGA ...................(((((((((..(.............)..))))))....)))(((((((((....)))))))))(((.....))).. (-18.81 = -18.71 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:28 2011