
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,817,776 – 2,817,869 |
| Length | 93 |
| Max. P | 0.562764 |

| Location | 2,817,776 – 2,817,869 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.03 |
| Shannon entropy | 0.38240 |
| G+C content | 0.66587 |
| Mean single sequence MFE | -40.55 |
| Consensus MFE | -26.40 |
| Energy contribution | -27.20 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.562764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2817776 93 + 21146708 GGGUCACCGACGAACUGCGUCUGACUGGCGGCGGUGGCUUGGACUGAUUAAUGGACGAUCGACGCCCGAUGCGCGUGGAGCCACCGCCGGCUG------------ .(((((..((((.....)))))))))(.(((((((((((((..(........)..)))((.((((.......)))).)))))))))))).)..------------ ( -43.60, z-score = -2.88, R) >droSim1.chr2R 1634254 93 + 19596830 GGGUCACCGACGAACUGCGUCUGACUGGCGGCGGUGGCUUGGCCUGAUUGAUAGACGAGCGACGCCCGAUGCGCGUGGAGCCACCGCCGGCUG------------ .(((((..((((.....)))))))))(.((((((((((((.((....(((.....)))(((.((.....))))))).)))))))))))).)..------------ ( -42.70, z-score = -1.98, R) >droSec1.super_1 499855 93 + 14215200 GGGUCACCGACGAACUGCGUCUGACUGGCGGCGGUGGCUUGGCCUGAUUGAUGGACGAGCGACGCCCGAUGCGCGUGGAGCCACCGCCGGCUG------------ .(((((..((((.....)))))))))(.((((((((((((.((.((((((..((.((.....)))))))).)).)).)))))))))))).)..------------ ( -44.90, z-score = -2.39, R) >droYak2.chr2L 15551058 93 + 22324452 GGGUCACCGACGAACUGCGUCUAACUGGCGGCGGUGGCUUGGCCUGAUUGAUGGACGCGCGACGGCCGAUGCGCGUGGAGCCACCGCUGGCUG------------ ((....))((((.....)))).....(.((((((((((((..((........))(((((((.(....).))))))).)))))))))))).)..------------ ( -44.40, z-score = -2.26, R) >droEre2.scaffold_4929 6909155 93 - 26641161 GGGUCACCGACGAACUGCGUCUAACUGGCGGCGGUGGCUUGGCCUGGUUGAUGGACGAGCGCCGUCCGAUGCGCGUGGAGCCACCGCCGGCUG------------ ((....))((((.....)))).....(.((((((((((((.((...((...((((((.....))))))..))..)).)))))))))))).)..------------ ( -46.20, z-score = -1.85, R) >droAna3.scaffold_13266 9143744 96 + 19884421 GGGUCACCGAAGAGCUGCGUCUGACUGGCGGCGGUGGCUUGGCCUGGUUGAUGGACGAACGGCGGGCGAUGCGCGUGGAUCCACCGCCACCUGCUC--------- ((....))...((((...(((.....)))((((((((.(((.((........)).))).(.(((.((...)).))).)..))))))))....))))--------- ( -36.10, z-score = 1.19, R) >droWil1.scaffold_180700 2337012 105 + 6630534 GUGUGACUGAUGAACUGCGUCUAACUGGUGGCGGUGGUUUUGCCUGAUUGAUGGAUGAUCGUCGUGCUAUGCGCGUUGAACCACCGCCCAUUGCUCCUCCGCCUC (((.((..((((.....))))......(.(((((((((((.((......((((......))))((((...)))))).))))))))))))........)))))... ( -33.40, z-score = -1.59, R) >droVir3.scaffold_12823 824357 93 - 2474545 GCGUCACCGACGAGCUGCGCCGCACUGGCGGCGGCGGCUUGGCCUGGUUAAUGGAAGCACGCCGUGCGAUGCGCGUUGAGCCACCUCCUGCUC------------ (((((((((.(((((((((((((....))))).))))))))...))))........((((...))))))))).....((((........))))------------ ( -41.20, z-score = -0.11, R) >droMoj3.scaffold_6496 25935171 96 - 26866924 GCGUCACUGAGGAGCUGCGUCUCACUGGCGGCGGGGGCUUGGCCUGAUUGAUGGAAGAACGACGCGCAAUGCGUGUGGAGCCAACUCCUGCCGCUC--------- ((((((.(((((.......))))).))))((((((((.(((((((..........))..(.((((((...)))))).).)))))))))))))))..--------- ( -41.70, z-score = -1.43, R) >droGri2.scaffold_15245 16381351 93 + 18325388 GCGUCACCGAUGAGCUGCGACGCACUGGCGGCGGUGGCUUCGCCUGAUUAAUGGAUGAACGACGUGCGAUACGUGUGGAGGAACUGCCCCCAC------------ (((((.(.(.....).).)))))..(((.((((((..((((((.(((((.(((.........)))..))).)).))))))..)))))).))).------------ ( -31.30, z-score = -0.18, R) >consensus GGGUCACCGACGAACUGCGUCUGACUGGCGGCGGUGGCUUGGCCUGAUUGAUGGACGAACGACGCCCGAUGCGCGUGGAGCCACCGCCGGCUG____________ ..((((..((((.....))))....))))(((((((((((..((........))....(((.(((.....)))))).)))))))))))................. (-26.40 = -27.20 + 0.80)

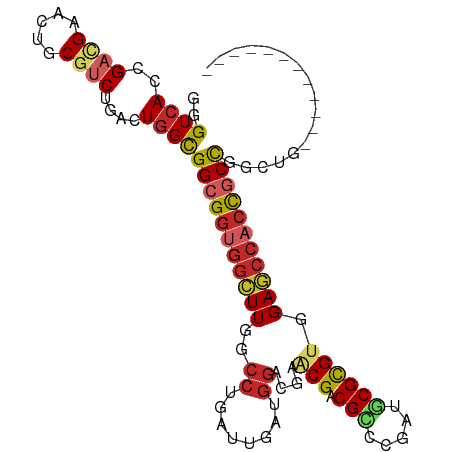
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:25 2011