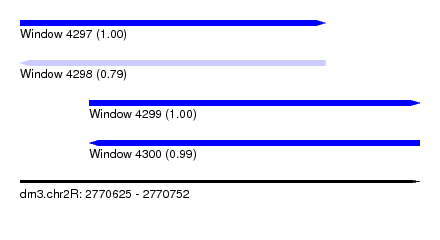
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,770,625 – 2,770,752 |
| Length | 127 |
| Max. P | 0.996385 |

| Location | 2,770,625 – 2,770,722 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.13 |
| Shannon entropy | 0.49807 |
| G+C content | 0.45714 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -19.02 |
| Energy contribution | -18.17 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.995784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
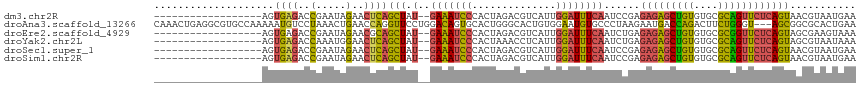
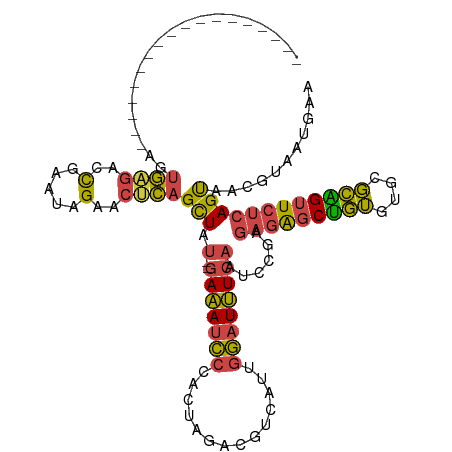
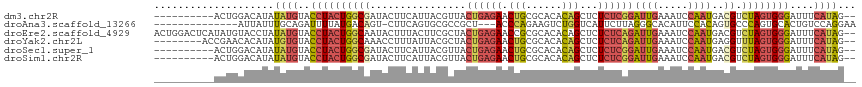
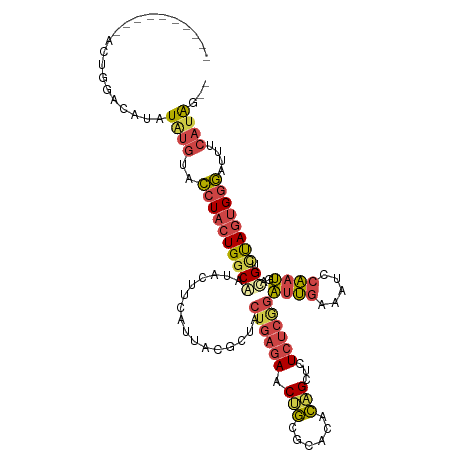
>dm3.chr2R 2770625 97 + 21146708 ------------------AGUGAGACCGAAUAGAACUCAGCUAU--GAAAUCCCACUAGACGUCAUUGGAUUUCAAUCCGAGAGAGCUGUGUGCGCAGUUCUCAGUAACGUAAUGAA ------------------((((.((....((((.......))))--....)).))))..((((..((((((....))))))(((((((((....)))))))))....))))...... ( -27.70, z-score = -2.23, R) >droAna3.scaffold_13266 9098982 114 + 19884421 CAAACUGAGGCGUGCCAAAAAUGUCCUAAACUGAACCAGGUUCCUGGACAGUGCACUGGGCACUGUGGAAUGUGCCCUAAGAAUGACCAGACUUCUGGGU---AGCGGCGCACUGAA ........((.(((((.............((((..((((....)))).))))((...((((((........)))))).........((((....))))..---.))))))).))... ( -40.40, z-score = -2.11, R) >droEre2.scaffold_4929 6863919 97 - 26641161 ------------------AGUGAGACCGAAUAGAACGCAGCUAU--GAAAUCCCACUAGACGUCAUUGGAUUUCAAUCUGAGAGAGCUGUGUGCGCGGUUCUCAGUAGCGAAGUAAA ------------------(.((((((((....(.((((((((.(--(((((((.((.....))....))))))))...(....))))))))).).))).))))).)........... ( -29.10, z-score = -1.87, R) >droYak2.chr2L 15504939 97 + 22324452 ------------------AGUGAGACCAAAUGGAACUCAGCUAU--GAAAUCCCACUAAACCUCAUUGGAUUUCAAUCUGAGAGAGCUGUGUGCGCAGUUCUCAGUAGCGUAAUAAA ------------------..((((.((....))..))))(((((--(((((((..............))))))).......(((((((((....))))))))).)))))........ ( -31.04, z-score = -3.52, R) >droSec1.super_1 452421 97 + 14215200 ------------------AGUGAGACCGAAUAGAACUCAGCUAU--GAAAUCCCACUAGACGUCAUUGGAUUUCAAUCCGAGAGAGCUGUGUGCGCAGUUCUCAGUAACGUAAUGAA ------------------((((.((....((((.......))))--....)).))))..((((..((((((....))))))(((((((((....)))))))))....))))...... ( -27.70, z-score = -2.23, R) >droSim1.chr2R 1583779 97 + 19596830 ------------------AGUGAGACCGAAUAGAACUCAGCUAU--GAAAUCCCACUAGACGUCAUUGGAUUUCAAUCCGAGAGAGCUGUGUGCGCAGUUCUCAGUAACGUAAUGAA ------------------((((.((....((((.......))))--....)).))))..((((..((((((....))))))(((((((((....)))))))))....))))...... ( -27.70, z-score = -2.23, R) >consensus __________________AGUGAGACCGAAUAGAACUCAGCUAU__GAAAUCCCACUAGACGUCAUUGGAUUUCAAUCCGAGAGAGCUGUGUGCGCAGUUCUCAGUAACGUAAUGAA ....................((((..(.....)..))))(((....(((((((..............))))))).......(((((((((....))))))))))))........... (-19.02 = -18.17 + -0.85)
| Location | 2,770,625 – 2,770,722 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.13 |
| Shannon entropy | 0.49807 |
| G+C content | 0.45714 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -14.21 |
| Energy contribution | -14.13 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2770625 97 - 21146708 UUCAUUACGUUACUGAGAACUGCGCACACAGCUCUCUCGGAUUGAAAUCCAAUGACGUCUAGUGGGAUUUC--AUAGCUGAGUUCUAUUCGGUCUCACU------------------ ......((((((..((((.(((......))).))))..((((....))))..))))))..(((((((((..--((((.......))))..)))))))))------------------ ( -25.30, z-score = -1.29, R) >droAna3.scaffold_13266 9098982 114 - 19884421 UUCAGUGCGCCGCU---ACCCAGAAGUCUGGUCAUUCUUAGGGCACAUUCCACAGUGCCCAGUGCACUGUCCAGGAACCUGGUUCAGUUUAGGACAUUUUUGGCACGCCUCAGUUUG ....(((.(((...---..((((....))))....(((((((((((........)))))).....((((.((((....))))..)))).))))).......))).)))......... ( -38.50, z-score = -2.21, R) >droEre2.scaffold_4929 6863919 97 + 26641161 UUUACUUCGCUACUGAGAACCGCGCACACAGCUCUCUCAGAUUGAAAUCCAAUGACGUCUAGUGGGAUUUC--AUAGCUGCGUUCUAUUCGGUCUCACU------------------ .............(((((.(((.(.((.(((((.........((((((((....((.....)).)))))))--).))))).)).)....))))))))..------------------ ( -23.90, z-score = -1.18, R) >droYak2.chr2L 15504939 97 - 22324452 UUUAUUACGCUACUGAGAACUGCGCACACAGCUCUCUCAGAUUGAAAUCCAAUGAGGUUUAGUGGGAUUUC--AUAGCUGAGUUCCAUUUGGUCUCACU------------------ ........((((((((((...((.......))..))))))..((((((((.((........)).)))))))--)))))((((..((....)).))))..------------------ ( -28.00, z-score = -2.11, R) >droSec1.super_1 452421 97 - 14215200 UUCAUUACGUUACUGAGAACUGCGCACACAGCUCUCUCGGAUUGAAAUCCAAUGACGUCUAGUGGGAUUUC--AUAGCUGAGUUCUAUUCGGUCUCACU------------------ ......((((((..((((.(((......))).))))..((((....))))..))))))..(((((((((..--((((.......))))..)))))))))------------------ ( -25.30, z-score = -1.29, R) >droSim1.chr2R 1583779 97 - 19596830 UUCAUUACGUUACUGAGAACUGCGCACACAGCUCUCUCGGAUUGAAAUCCAAUGACGUCUAGUGGGAUUUC--AUAGCUGAGUUCUAUUCGGUCUCACU------------------ ......((((((..((((.(((......))).))))..((((....))))..))))))..(((((((((..--((((.......))))..)))))))))------------------ ( -25.30, z-score = -1.29, R) >consensus UUCAUUACGCUACUGAGAACUGCGCACACAGCUCUCUCGGAUUGAAAUCCAAUGACGUCUAGUGGGAUUUC__AUAGCUGAGUUCUAUUCGGUCUCACU__________________ .............((((.((((.((.....))...(((((...(((((((.((........)).)))))))......))))).......)))))))).................... (-14.21 = -14.13 + -0.07)

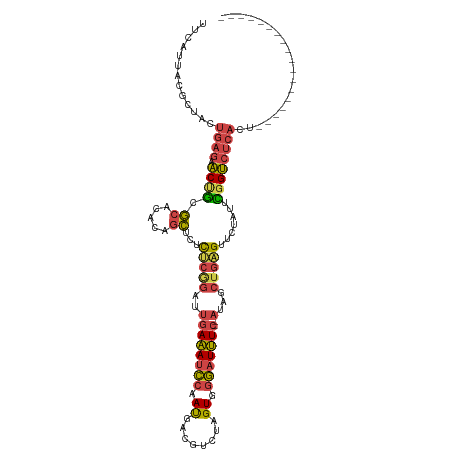
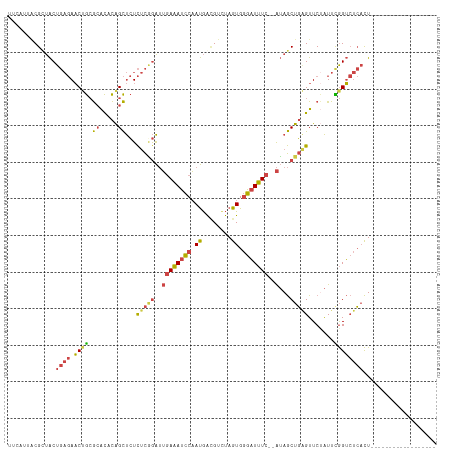
| Location | 2,770,647 – 2,770,752 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 70.12 |
| Shannon entropy | 0.55133 |
| G+C content | 0.44397 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -20.50 |
| Energy contribution | -19.92 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.996385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
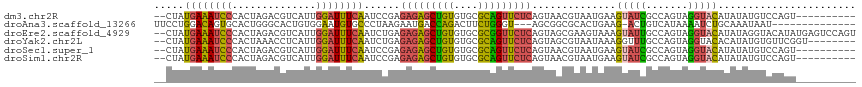
>dm3.chr2R 2770647 105 + 21146708 --CUAUGAAAUCCCACUAGACGUCAUUGGAUUUCAAUCCGAGAGAGCUGUGUGCGCAGUUCUCAGUAACGUAAUGAAGUAUCGCCAGUAGGUACAUAUAUGUCCAGU---------- --............(((.(((((((((((((....))))..(((((((((....)))))))))........))))).(((((.......))))).....)))).)))---------- ( -29.60, z-score = -2.59, R) >droAna3.scaffold_13266 9099022 99 + 19884421 UUCCUGGACAGUGCACUGGGCACUGUGGAAUGUGCCCUAAGAAUGACCAGACUUCUGGGU---AGCGGCGCACUGAAG-ACUGUCAUAAAAUCUGCAAAUAAU-------------- ........((((((.((((((((........)))))).........((((....))))..---...)).))))))...-........................-------------- ( -29.60, z-score = -1.39, R) >droEre2.scaffold_4929 6863941 115 - 26641161 --CUAUGAAAUCCCACUAGACGUCAUUGGAUUUCAAUCUGAGAGAGCUGUGUGCGCGGUUCUCAGUAGCGAAGUAAAGUAUUGCCAGUAGGUACAUAUAGGUACAUAUGAGUCCAGU --...((((((((.((.....))....))))))))..(((.(((((((((....))))))))).........(((..((((((((....)))).))))...))).........))). ( -29.30, z-score = -0.86, R) >droYak2.chr2L 15504961 107 + 22324452 --CUAUGAAAUCCCACUAAACCUCAUUGGAUUUCAAUCUGAGAGAGCUGUGUGCGCAGUUCUCAGUAGCGUAAUAAAGGUUUGCCAGUAGGUACACAUAUGUGUUCGGU-------- --...((((((((..............))))))))..(((((((((((((....)))))))))....(((((......((.((((....)))).)).)))))..)))).-------- ( -30.34, z-score = -1.93, R) >droSec1.super_1 452443 105 + 14215200 --CUAUGAAAUCCCACUAGACGUCAUUGGAUUUCAAUCCGAGAGAGCUGUGUGCGCAGUUCUCAGUAACGUAAUGAAGUAUCGCCAGUAGGUACAUAUAUGUCCAGU---------- --............(((.(((((((((((((....))))..(((((((((....)))))))))........))))).(((((.......))))).....)))).)))---------- ( -29.60, z-score = -2.59, R) >droSim1.chr2R 1583801 105 + 19596830 --CUAUGAAAUCCCACUAGACGUCAUUGGAUUUCAAUCCGAGAGAGCUGUGUGCGCAGUUCUCAGUAACGUAAUGAAGUAUCGCCAGUAGGUACAUAUAUGUCCAGU---------- --............(((.(((((((((((((....))))..(((((((((....)))))))))........))))).(((((.......))))).....)))).)))---------- ( -29.60, z-score = -2.59, R) >consensus __CUAUGAAAUCCCACUAGACGUCAUUGGAUUUCAAUCCGAGAGAGCUGUGUGCGCAGUUCUCAGUAACGUAAUGAAGUAUCGCCAGUAGGUACAUAUAUGUCCAGU__________ .....((((((((..............))))))))......(((((((((....)))))))))..............(((((.......)))))....................... (-20.50 = -19.92 + -0.57)
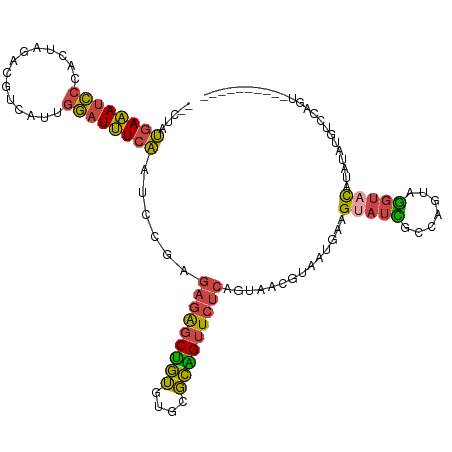
| Location | 2,770,647 – 2,770,752 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.12 |
| Shannon entropy | 0.55133 |
| G+C content | 0.44397 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -15.16 |
| Energy contribution | -15.83 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2770647 105 - 21146708 ----------ACUGGACAUAUAUGUACCUACUGGCGAUACUUCAUUACGUUACUGAGAACUGCGCACACAGCUCUCUCGGAUUGAAAUCCAAUGACGUCUAGUGGGAUUUCAUAG-- ----------..........((((..((((((((.((....))...((((((..((((.(((......))).))))..((((....))))..))))))))))))))....)))).-- ( -27.40, z-score = -2.12, R) >droAna3.scaffold_13266 9099022 99 - 19884421 --------------AUUAUUUGCAGAUUUUAUGACAGU-CUUCAGUGCGCCGCU---ACCCAGAAGUCUGGUCAUUCUUAGGGCACAUUCCACAGUGCCCAGUGCACUGUCCAGGAA --------------.......................(-(((((((((((....---..((((....)))).........((((((........)))))).))))))))...)))). ( -31.10, z-score = -2.40, R) >droEre2.scaffold_4929 6863941 115 + 26641161 ACUGGACUCAUAUGUACCUAUAUGUACCUACUGGCAAUACUUUACUUCGCUACUGAGAACCGCGCACACAGCUCUCUCAGAUUGAAAUCCAAUGACGUCUAGUGGGAUUUCAUAG-- ...((((......)).))..((((..(((((((((.................((((((...((.......))..))))))((((.....))))...).))))))))....)))).-- ( -25.50, z-score = -1.25, R) >droYak2.chr2L 15504961 107 - 22324452 --------ACCGAACACAUAUGUGUACCUACUGGCAAACCUUUAUUACGCUACUGAGAACUGCGCACACAGCUCUCUCAGAUUGAAAUCCAAUGAGGUUUAGUGGGAUUUCAUAG-- --------...((((((....)))).(((((((...((((((..........((((((...((.......))..))))))((((.....)))))))))))))))))...))....-- ( -28.30, z-score = -2.47, R) >droSec1.super_1 452443 105 - 14215200 ----------ACUGGACAUAUAUGUACCUACUGGCGAUACUUCAUUACGUUACUGAGAACUGCGCACACAGCUCUCUCGGAUUGAAAUCCAAUGACGUCUAGUGGGAUUUCAUAG-- ----------..........((((..((((((((.((....))...((((((..((((.(((......))).))))..((((....))))..))))))))))))))....)))).-- ( -27.40, z-score = -2.12, R) >droSim1.chr2R 1583801 105 - 19596830 ----------ACUGGACAUAUAUGUACCUACUGGCGAUACUUCAUUACGUUACUGAGAACUGCGCACACAGCUCUCUCGGAUUGAAAUCCAAUGACGUCUAGUGGGAUUUCAUAG-- ----------..........((((..((((((((.((....))...((((((..((((.(((......))).))))..((((....))))..))))))))))))))....)))).-- ( -27.40, z-score = -2.12, R) >consensus __________ACUGGACAUAUAUGUACCUACUGGCAAUACUUCAUUACGCUACUGAGAACUGCGCACACAGCUCUCUCGGAUUGAAAUCCAAUGACGUCUAGUGGGAUUUCAUAG__ ....................((((..((((((((((................((((((.(((......)))...))))))((((.....))))..)).))))))))....))))... (-15.16 = -15.83 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:16 2011