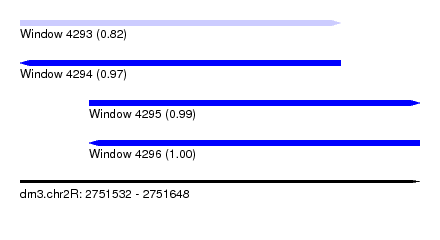
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,751,532 – 2,751,648 |
| Length | 116 |
| Max. P | 0.995582 |

| Location | 2,751,532 – 2,751,625 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.53 |
| Shannon entropy | 0.42965 |
| G+C content | 0.57053 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -16.25 |
| Energy contribution | -15.72 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
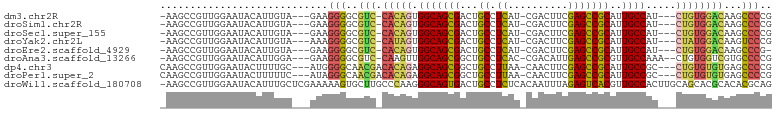
>dm3.chr2R 2751532 93 + 21146708 -AAGCCGUUGGAAUACAUUGUA---GAAGGGGCGUC-CACAGUGGCAGCGACUGCCUCAU-CGACUUCGAGCCGCAUUGCCAU---CUGUGGACAAGCCCCG -.....................---...((((((((-(((((((((((....(((....(-((....)))...))))))))).---))))))))..))))). ( -38.60, z-score = -3.05, R) >droSim1.chr2R 1564281 93 + 19596830 -AAGCCGUUGGAAUACAUUGUA---GAAGGGGCGUC-CACAGUGGCAGCGACUGCCUCAU-CGACUUCGAGCCGCAUUGCCAU---CUGUGGACAAGCCCCG -.....................---...((((((((-(((((((((((....(((....(-((....)))...))))))))).---))))))))..))))). ( -38.60, z-score = -3.05, R) >droSec1.super_155 28009 93 + 49466 -AAGCCGUUGGAAUACAUUGUA---GAAGGGGCGUC-CACAGUGGCAGCGACUGCCUCAU-CGACUUCGAGCCGCAUUGCCAU---CUGUGGACAAGCCCCG -.....................---...((((((((-(((((((((((....(((....(-((....)))...))))))))).---))))))))..))))). ( -38.60, z-score = -3.05, R) >droYak2.chr2L 15484810 93 + 22324452 -AAGCCGUUGGAAUACAUUGUA---AAAGGGGCGUC-CAUAGUGGCAGCGACUGCCUCAU-CGACUUCGAGCCGCAUUGCCAU---CUAUGGACAAGUCCCG -.....................---...((((((((-(((((((((((....(((....(-((....)))...))))))))).---))))))))..))))). ( -34.00, z-score = -2.60, R) >droEre2.scaffold_4929 6844229 92 - 26641161 -AAGCCGUUGGAAUACAUUGUA---GAAGGGGCGUC-CACAGUGGCAGCGACUGCCUCAU-CGACUUCGAGCCGCAUUGCCAU---CUGUGGACAAGCCCG- -.....................---....(((((((-(((((((((((....(((....(-((....)))...))))))))).---))))))))..)))).- ( -35.10, z-score = -2.07, R) >droAna3.scaffold_13266 9081621 94 + 19884421 -AAGCCGUUGGAAUACAUUGGA---GAAGGGGCGUC-CAAGUUGGCAGCGGCUGCCUCAC-CGACAUUGAGCCGCGUUGCCAAA--CUGUGGUCGUGCCCCG -...(((.((.....)).))).---...((((((.(-((..(((((((((.(.((.(((.-......))))).)))))))))).--...)))...)))))). ( -37.30, z-score = -1.25, R) >dp4.chr3 1020920 95 + 19779522 CAAGCCGUUGGAAUACUUUUGC---AUGGGGCAACGACACAGAGGCAGCGGCUGCCUUAA-CAACUUCGAGCCGCAUUGCCGC---CUGUGUGUGAGCCCCG ...((.(..(.....)..).))---..(((((.((.((((((.((((((((((.......-........))))))..))))..---))))))))..))))). ( -38.56, z-score = -2.27, R) >droPer1.super_2 1201817 95 + 9036312 CAAGCCGUUGGAAUACUUUUUC---AUAGGGCAACGACACAGAGGCAGCGGCUGCCUUAA-CAACUUCGAGCCGCAUUGCCGC---CUGUGUGUGAGCCCCG ........(((((.....))))---)..((((.((.((((((.((((((((((.......-........))))))..))))..---))))))))..)))).. ( -35.26, z-score = -2.28, R) >droWil1.scaffold_180708 76870 101 - 12563649 -AAGCCGUUGGAAUACAUUUGCUCGAAAAAGUGCUUGCCCAAGGGCAGUGACUGCCUCUCACAAUUUAGAGUCACGUUGCCACUUGCAGCACGCACACGCAG -.(((...((.....))...))).......((((.(((.((((((((((((((................))))))..)))).))))..))).))))...... ( -28.89, z-score = -0.46, R) >consensus _AAGCCGUUGGAAUACAUUGUA___GAAGGGGCGUC_CACAGUGGCAGCGACUGCCUCAU_CGACUUCGAGCCGCAUUGCCAU___CUGUGGACAAGCCCCG ............................(((..(((.(((((.(((((((...((.((..........)))))))..)))).....))))))))...))).. (-16.25 = -15.72 + -0.52)

| Location | 2,751,532 – 2,751,625 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.53 |
| Shannon entropy | 0.42965 |
| G+C content | 0.57053 |
| Mean single sequence MFE | -38.06 |
| Consensus MFE | -20.88 |
| Energy contribution | -20.72 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2751532 93 - 21146708 CGGGGCUUGUCCACAG---AUGGCAAUGCGGCUCGAAGUCG-AUGAGGCAGUCGCUGCCACUGUG-GACGCCCCUUC---UACAAUGUAUUCCAACGGCUU- .(((((..((((((((---.(((((..((((((....(((.-....)))))))))))))))))))-))))))))...---.....................- ( -42.80, z-score = -3.72, R) >droSim1.chr2R 1564281 93 - 19596830 CGGGGCUUGUCCACAG---AUGGCAAUGCGGCUCGAAGUCG-AUGAGGCAGUCGCUGCCACUGUG-GACGCCCCUUC---UACAAUGUAUUCCAACGGCUU- .(((((..((((((((---.(((((..((((((....(((.-....)))))))))))))))))))-))))))))...---.....................- ( -42.80, z-score = -3.72, R) >droSec1.super_155 28009 93 - 49466 CGGGGCUUGUCCACAG---AUGGCAAUGCGGCUCGAAGUCG-AUGAGGCAGUCGCUGCCACUGUG-GACGCCCCUUC---UACAAUGUAUUCCAACGGCUU- .(((((..((((((((---.(((((..((((((....(((.-....)))))))))))))))))))-))))))))...---.....................- ( -42.80, z-score = -3.72, R) >droYak2.chr2L 15484810 93 - 22324452 CGGGACUUGUCCAUAG---AUGGCAAUGCGGCUCGAAGUCG-AUGAGGCAGUCGCUGCCACUAUG-GACGCCCCUUU---UACAAUGUAUUCCAACGGCUU- .(((...(((((((((---.(((((..((((((....(((.-....)))))))))))))))))))-)))).)))...---.....................- ( -35.00, z-score = -2.57, R) >droEre2.scaffold_4929 6844229 92 + 26641161 -CGGGCUUGUCCACAG---AUGGCAAUGCGGCUCGAAGUCG-AUGAGGCAGUCGCUGCCACUGUG-GACGCCCCUUC---UACAAUGUAUUCCAACGGCUU- -.((((..((((((((---.(((((..((((((....(((.-....)))))))))))))))))))-)))))))....---.....................- ( -39.10, z-score = -2.99, R) >droAna3.scaffold_13266 9081621 94 - 19884421 CGGGGCACGACCACAG--UUUGGCAACGCGGCUCAAUGUCG-GUGAGGCAGCCGCUGCCAACUUG-GACGCCCCUUC---UCCAAUGUAUUCCAACGGCUU- .(((((....(((.((--((.((((..((((((....(((.-....)))))))))))))))))))-)..)))))...---.....................- ( -34.40, z-score = -0.96, R) >dp4.chr3 1020920 95 - 19779522 CGGGGCUCACACACAG---GCGGCAAUGCGGCUCGAAGUUG-UUAAGGCAGCCGCUGCCUCUGUGUCGUUGCCCCAU---GCAAAAGUAUUCCAACGGCUUG .(((((..((((((((---(.((((..((((((....(((.-....)))))))))))))))))))).)).)))))..---((....((......)).))... ( -38.80, z-score = -1.91, R) >droPer1.super_2 1201817 95 - 9036312 CGGGGCUCACACACAG---GCGGCAAUGCGGCUCGAAGUUG-UUAAGGCAGCCGCUGCCUCUGUGUCGUUGCCCUAU---GAAAAAGUAUUCCAACGGCUUG .(((((..((((((((---(.((((..((((((....(((.-....)))))))))))))))))))).)).)))))..---....((((.........)))). ( -35.50, z-score = -1.47, R) >droWil1.scaffold_180708 76870 101 + 12563649 CUGCGUGUGCGUGCUGCAAGUGGCAACGUGACUCUAAAUUGUGAGAGGCAGUCACUGCCCUUGGGCAAGCACUUUUUCGAGCAAAUGUAUUCCAACGGCUU- ......((((.((((.((((.((((..((((((((...........)).)))))))))))))))))).))))......((((...((.....))...))))- ( -31.30, z-score = 0.10, R) >consensus CGGGGCUUGUCCACAG___AUGGCAAUGCGGCUCGAAGUCG_AUGAGGCAGUCGCUGCCACUGUG_GACGCCCCUUC___UACAAUGUAUUCCAACGGCUU_ .((((......(((((.....((((..((((((....(((......))))))))))))).)))))......))))........................... (-20.88 = -20.72 + -0.15)

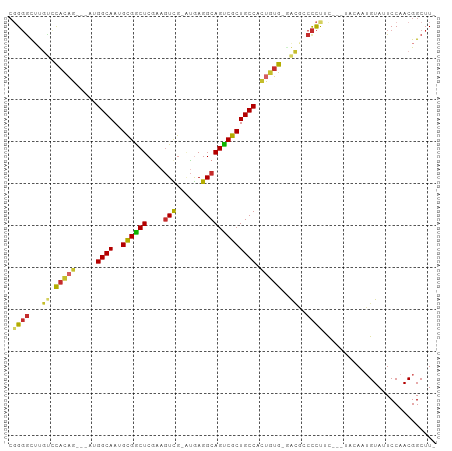
| Location | 2,751,552 – 2,751,648 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.20 |
| Shannon entropy | 0.56171 |
| G+C content | 0.56398 |
| Mean single sequence MFE | -38.87 |
| Consensus MFE | -16.91 |
| Energy contribution | -17.04 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
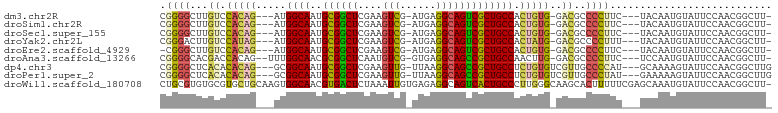
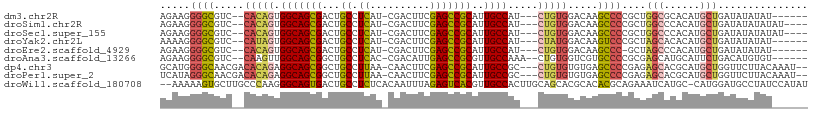
>dm3.chr2R 2751552 96 + 21146708 AGAAGGGGCGUC--CACAGUGGCAGCGACUGCCUCAU-CGACUUCGAGCCGCAUUGCCAU---CUGUGGACAAGCCCCGCUGGCGCACAUGCUGAUAUAUAU------ ....((((((((--(((((((((((....(((....(-((....)))...))))))))).---))))))))..)))))((((.....)).))..........------ ( -40.70, z-score = -2.60, R) >droSim1.chr2R 1564301 98 + 19596830 AGAAGGGGCGUC--CACAGUGGCAGCGACUGCCUCAU-CGACUUCGAGCCGCAUUGCCAU---CUGUGGACAAGCCCCGCUGGCCCACAUGCUGAUAUAUAUAU---- ....((((((((--(((((((((((....(((....(-((....)))...))))))))).---))))))))..)))))((((.....)).))............---- ( -40.70, z-score = -2.95, R) >droSec1.super_155 28029 98 + 49466 AGAAGGGGCGUC--CACAGUGGCAGCGACUGCCUCAU-CGACUUCGAGCCGCAUUGCCAU---CUGUGGACAAGCCCCGCUGGCCCACAUGCUGAUAUAUAUAU---- ....((((((((--(((((((((((....(((....(-((....)))...))))))))).---))))))))..)))))((((.....)).))............---- ( -40.70, z-score = -2.95, R) >droYak2.chr2L 15484830 96 + 22324452 AAAAGGGGCGUC--CAUAGUGGCAGCGACUGCCUCAU-CGACUUCGAGCCGCAUUGCCAU---CUAUGGACAAGUCCCGCUAGCACACAUGCUGAUAUAUAU------ ....((((((((--(((((((((((....(((....(-((....)))...))))))))).---))))))))..)))))..(((((....)))))........------ ( -37.50, z-score = -3.51, R) >droEre2.scaffold_4929 6844249 95 - 26641161 AGAAGGGGCGUC--CACAGUGGCAGCGACUGCCUCAU-CGACUUCGAGCCGCAUUGCCAU---CUGUGGACAAGCCC-GCUAGCCCACAUGCUGAUAUAUAU------ .....(((((((--(((((((((((....(((....(-((....)))...))))))))).---))))))))..))))-..((((......))))........------ ( -37.40, z-score = -2.53, R) >droAna3.scaffold_13266 9081641 97 + 19884421 AGAAGGGGCGUC--CAAGUUGGCAGCGGCUGCCUCAC-CGACAUUGAGCCGCGUUGCCAAA--CUGUGGUCGUGCCCCGCGAGCAUGCAUUCUGACAUGUGU------ ....((((((.(--((..(((((((((.(.((.(((.-......))))).)))))))))).--...)))...))))))....((((((.....).)))))..------ ( -38.20, z-score = -0.75, R) >dp4.chr3 1020940 102 + 19779522 GCAUGGGGCAACGACACAGAGGCAGCGGCUGCCUUAA-CAACUUCGAGCCGCAUUGCCGC---CUGUGUGUGAGCCCCGAGAGCACGCAUGCUGGUUCUUACAAAU-- ....(((((.((.((((((.((((((((((.......-........))))))..))))..---))))))))..)))))((((((.((.....))))))))......-- ( -42.56, z-score = -2.38, R) >droPer1.super_2 1201837 102 + 9036312 UCAUAGGGCAACGACACAGAGGCAGCGGCUGCCUUAA-CAACUUCGAGCCGCAUUGCCGC---CUGUGUGUGAGCCCCGAGAGCACGCAUGCUGGUUCUUACAAAU-- .....((((.((.((((((.((((((((((.......-........))))))..))))..---))))))))..)))).((((((.((.....))))))))......-- ( -38.56, z-score = -1.97, R) >droWil1.scaffold_180708 76894 105 - 12563649 --AAAAAGUGCUUGCCCAAGGGCAGUGACUGCCUCUCACAAUUUAGAGUCACGUUGCCACUUGCAGCACGCACACGCAGAAAUCAUGC-CAUGGAUGCCUAUCCAUAU --.....((((.(((.((((((((((((((................))))))..)))).))))..))).))))..(((.......)))-.((((((....)))))).. ( -33.49, z-score = -2.25, R) >consensus AGAAGGGGCGUC__CACAGUGGCAGCGACUGCCUCAU_CGACUUCGAGCCGCAUUGCCAU___CUGUGGACAAGCCCCGCUAGCACACAUGCUGAUAUAUAU______ .....((((.....(((((.(((((((...((.((..........)))))))..)))).....))))).....))))....(((......)))............... (-16.91 = -17.04 + 0.13)
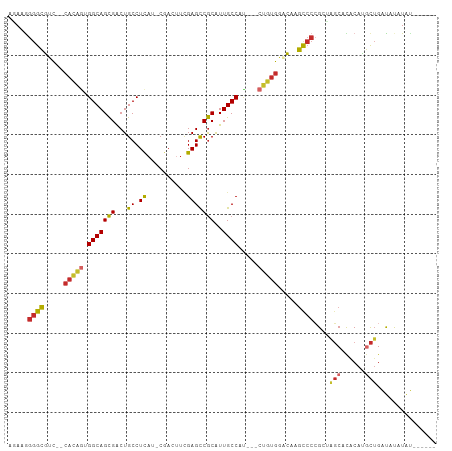
| Location | 2,751,552 – 2,751,648 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 72.20 |
| Shannon entropy | 0.56171 |
| G+C content | 0.56398 |
| Mean single sequence MFE | -40.61 |
| Consensus MFE | -25.90 |
| Energy contribution | -25.54 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
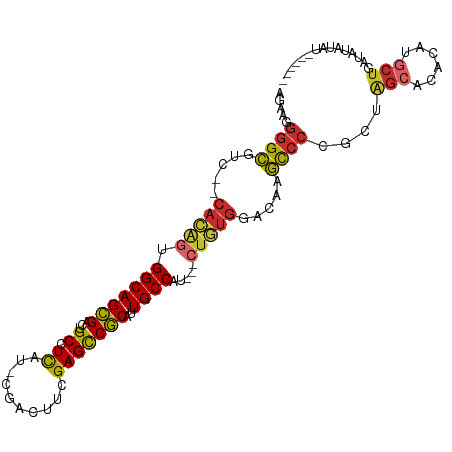
>dm3.chr2R 2751552 96 - 21146708 ------AUAUAUAUCAGCAUGUGCGCCAGCGGGGCUUGUCCACAG---AUGGCAAUGCGGCUCGAAGUCG-AUGAGGCAGUCGCUGCCACUGUG--GACGCCCCUUCU ------................((....))(((((..((((((((---.(((((..((((((....(((.-....)))))))))))))))))))--)))))))).... ( -44.50, z-score = -2.98, R) >droSim1.chr2R 1564301 98 - 19596830 ----AUAUAUAUAUCAGCAUGUGGGCCAGCGGGGCUUGUCCACAG---AUGGCAAUGCGGCUCGAAGUCG-AUGAGGCAGUCGCUGCCACUGUG--GACGCCCCUUCU ----............((.((.....))))(((((..((((((((---.(((((..((((((....(((.-....)))))))))))))))))))--)))))))).... ( -43.90, z-score = -2.67, R) >droSec1.super_155 28029 98 - 49466 ----AUAUAUAUAUCAGCAUGUGGGCCAGCGGGGCUUGUCCACAG---AUGGCAAUGCGGCUCGAAGUCG-AUGAGGCAGUCGCUGCCACUGUG--GACGCCCCUUCU ----............((.((.....))))(((((..((((((((---.(((((..((((((....(((.-....)))))))))))))))))))--)))))))).... ( -43.90, z-score = -2.67, R) >droYak2.chr2L 15484830 96 - 22324452 ------AUAUAUAUCAGCAUGUGUGCUAGCGGGACUUGUCCAUAG---AUGGCAAUGCGGCUCGAAGUCG-AUGAGGCAGUCGCUGCCACUAUG--GACGCCCCUUUU ------.........((((....))))...(((...(((((((((---.(((((..((((((....(((.-....)))))))))))))))))))--)))).))).... ( -38.10, z-score = -2.56, R) >droEre2.scaffold_4929 6844249 95 + 26641161 ------AUAUAUAUCAGCAUGUGGGCUAGC-GGGCUUGUCCACAG---AUGGCAAUGCGGCUCGAAGUCG-AUGAGGCAGUCGCUGCCACUGUG--GACGCCCCUUCU ------.........(((......)))((.-((((..((((((((---.(((((..((((((....(((.-....)))))))))))))))))))--)))))))))... ( -41.40, z-score = -2.43, R) >droAna3.scaffold_13266 9081641 97 - 19884421 ------ACACAUGUCAGAAUGCAUGCUCGCGGGGCACGACCACAG--UUUGGCAACGCGGCUCAAUGUCG-GUGAGGCAGCCGCUGCCAACUUG--GACGCCCCUUCU ------...(((((......))))).....(((((....(((.((--((.((((..((((((....(((.-....)))))))))))))))))))--)..))))).... ( -37.70, z-score = -1.15, R) >dp4.chr3 1020940 102 - 19779522 --AUUUGUAAGAACCAGCAUGCGUGCUCUCGGGGCUCACACACAG---GCGGCAAUGCGGCUCGAAGUUG-UUAAGGCAGCCGCUGCCUCUGUGUCGUUGCCCCAUGC --........((...((((....)))).))(((((..((((((((---(.((((..((((((....(((.-....)))))))))))))))))))).)).))))).... ( -40.80, z-score = -1.29, R) >droPer1.super_2 1201837 102 - 9036312 --AUUUGUAAGAACCAGCAUGCGUGCUCUCGGGGCUCACACACAG---GCGGCAAUGCGGCUCGAAGUUG-UUAAGGCAGCCGCUGCCUCUGUGUCGUUGCCCUAUGA --........((...((((....)))).))(((((..((((((((---(.((((..((((((....(((.-....)))))))))))))))))))).)).))))).... ( -38.10, z-score = -0.73, R) >droWil1.scaffold_180708 76894 105 + 12563649 AUAUGGAUAGGCAUCCAUG-GCAUGAUUUCUGCGUGUGCGUGCUGCAAGUGGCAACGUGACUCUAAAUUGUGAGAGGCAGUCACUGCCCUUGGGCAAGCACUUUUU-- .(((((((....)))))))-(((.(....))))..((((.((((.((((.((((..((((((((...........)).)))))))))))))))))).)))).....-- ( -37.10, z-score = -1.21, R) >consensus ______AUAUAUAUCAGCAUGUGUGCUAGCGGGGCUUGUCCACAG___AUGGCAAUGCGGCUCGAAGUCG_AUGAGGCAGUCGCUGCCACUGUG__GACGCCCCUUCU ..............................(((((.....(((((.....((((..((((((....(((......))))))))))))).))))).....))))).... (-25.90 = -25.54 + -0.35)
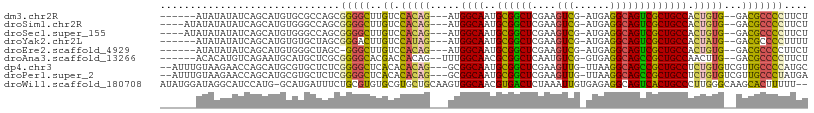

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:13 2011