| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,747,655 – 2,747,750 |
| Length | 95 |
| Max. P | 0.830940 |

| Location | 2,747,655 – 2,747,750 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.86 |
| Shannon entropy | 0.31666 |
| G+C content | 0.36013 |
| Mean single sequence MFE | -11.71 |
| Consensus MFE | -6.66 |
| Energy contribution | -6.88 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.830940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
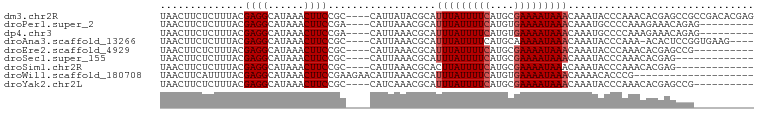
>dm3.chr2R 2747655 95 + 21146708 UAACUUCUCUUUACGAGGCAUAAACUUCCGC----CAUUAUACGCAUUUAUUUUCAUGCGAAAAUAAACAAAUACCCAAACACGAGCCGCCGACACGAG ......(((....((.(((..........((----........)).(((((((((....))))))))).................)))..))....))) ( -14.30, z-score = -2.18, R) >droPer1.super_2 1198697 86 + 9036312 UAACUUCUCUUUACGAGGCAUAAACUUCCGA----CAUUAAACGCAUUUAUUUUCAUGUGAAAAUAAACAAAUGCCCCAAAGAAACAGAG--------- .......(((((..(.(((((.......((.----.......))..(((((((((....)))))))))...)))))).))))).......--------- ( -13.40, z-score = -2.55, R) >dp4.chr3 1017796 86 + 19779522 UAACUUCUCUUUACGAGGCAUAAACUUCCGA----CAUUAAACGCAUUUAUUUUCAUGUGAAAAUAAACAAAUGCCCCAAAGAAACAGAG--------- .......(((((..(.(((((.......((.----.......))..(((((((((....)))))))))...)))))).))))).......--------- ( -13.40, z-score = -2.55, R) >droAna3.scaffold_13266 9078253 90 + 19884421 UAACUUCUCUUUACGAGGCAUAAACUUCCGC----CAUUAAACGCAUUUAUUUUCAUGCAAAAAUAAACAAAUACCCAAA-ACACUCCGGUGAAG---- ........((((((..(((..........))----).......((((........)))).....................-........))))))---- ( -11.40, z-score = -1.95, R) >droEre2.scaffold_4929 6840330 85 - 26641161 UAACUUCUCUUUACGAGGCAUAAACUUCCGC----CAUUAAACGCAUUUAUUUUCAUGCGAAAAUAAACAAAUACCCAAACACGAGCCG---------- ................(((..........((----........)).(((((((((....))))))))).................))).---------- ( -12.10, z-score = -2.59, R) >droSec1.super_155 24077 82 + 49466 UAACUUCUCUUUACGAGGCAUAAACUUCCGC----CAUUAAACGCAUUUAUUUUCAUGCGAAAAUAAACAAAUACCCAAACACGAG------------- ......(((.......(((..........))----)..........(((((((((....)))))))))...............)))------------- ( -10.60, z-score = -2.62, R) >droSim1.chr2R 1560404 82 + 19596830 UAACUUCUCUUUACGAGGCAUAAACUUCCGC----CAUUAAACGCACUUAUUUUCAUGCGAAAAUAAACAAAUACCCAAACACGAG------------- ......(((.......(((..........))----)...........((((((((....))))))))................)))------------- ( -9.80, z-score = -2.48, R) >droWil1.scaffold_180708 65167 79 - 12563649 UAACUUCAUUUUACGAGGCAUAAACUUCCGAAGAACAUUAAACGCAUUUAUUUUCAUGUGAAAAUAAACAAAACACCCG-------------------- ...((((.......((((......)))).)))).............(((((((((....)))))))))...........-------------------- ( -8.30, z-score = -1.31, R) >droYak2.chr2L 15481312 85 + 22324452 UAACUUCUCUUUACGAGGCAUAAACUUCCGC----CAUCAAACGCAUUUAUUUUCAUGCGAAAAUAAACAAAUACCCAAACACGAGCCG---------- ................(((..........((----........)).(((((((((....))))))))).................))).---------- ( -12.10, z-score = -2.62, R) >consensus UAACUUCUCUUUACGAGGCAUAAACUUCCGC____CAUUAAACGCAUUUAUUUUCAUGCGAAAAUAAACAAAUACCCAAACACGAGCCG__________ ..............((((......))))..................(((((((((....)))))))))............................... ( -6.66 = -6.88 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:10 2011