| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,710,329 – 2,710,424 |
| Length | 95 |
| Max. P | 0.926652 |

| Location | 2,710,329 – 2,710,424 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.58 |
| Shannon entropy | 0.51257 |
| G+C content | 0.45536 |
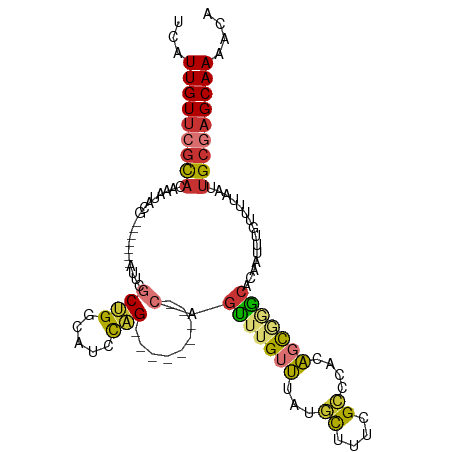
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -14.59 |
| Energy contribution | -14.30 |
| Covariance contribution | -0.29 |
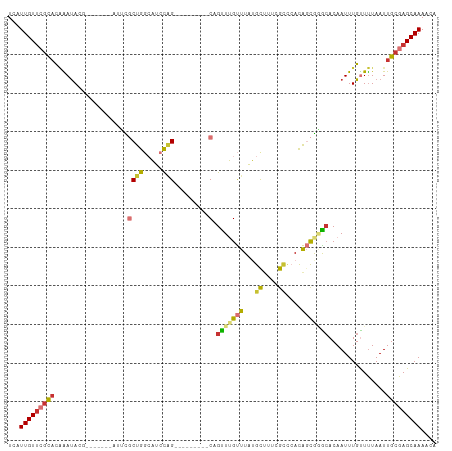
| Combinations/Pair | 1.52 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.926652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
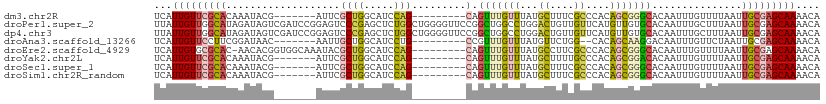
>dm3.chr2R 2710329 95 - 21146708 UCAUUGUUCGCACAAAUACG-------AUUCGCUGGCAUCCAG---------CAGUUUGUUUAUGCUUUCGCCCACAGCGGGCACAAUUUGUUUUAAUUGCGAGCAAAACA ...(((((((((.....(((-------(...(((((...))))---------).................((((.....)))).....))))......))))))))).... ( -28.90, z-score = -2.64, R) >droPer1.super_2 1163178 111 - 9036312 UUAUUGUUGGCAUAGAUAGUCGAUCCGGAGUCCCGAGCUCUGGCUGGGGUUCCGGCUGGCCUGGACUGUUGUUCAUGUUGUGCACAAUUUGCUUUAAUUGCGAGCAAAACA ......(((((((((...((((..(((((..(((.(((....))))))..))))).)))).(((((....)))))..)))))).)))(((((((.......)))))))... ( -39.90, z-score = -2.03, R) >dp4.chr3 984978 111 - 19779522 UUAUUGUUGGCAUAGAUAGUCGAUCCGGAGUCCCGAGCUCUGGCUGGGGUUCCGGCUGGCCUGGACUGUUGUUCAUGUUGUGCACAAUUUGCUUUAAUUGCGAGCAAAACA ......(((((((((...((((..(((((..(((.(((....))))))..))))).)))).(((((....)))))..)))))).)))(((((((.......)))))))... ( -39.90, z-score = -2.03, R) >droAna3.scaffold_13266 9038778 93 - 19884421 UCAUUGUUCCUUCGGAUAAC-------AAUUGCUGGCAUCCUG---------CCGUUUGUUUAUGUUCUGG--CACAGCAAGGACAAUUUGUUCUAAUUGCGAGCAAAACA ...((((((..(((((.(((-------((....((((.....)---------))).))))).....)))))--....((((((((.....)))))...))))))))).... ( -22.10, z-score = -0.54, R) >droEre2.scaffold_4929 6803406 101 + 26641161 UCAUUGUGCGCAC-AACACGGUGGCAAAUACGCUGGCAUCCAG---------CAGUUUGUUUAUGCCUUCGCCCACAGCGGGCACAAUUUGUUUUAAUUGCGAGCAAAACA ...((((.((((.-.....((((((((((..(((((...))))---------).)))))))...)))...((((.....))))...............)))).)))).... ( -30.00, z-score = -0.55, R) >droYak2.chr2L 15444726 95 - 22324452 UCAUUGUUCGCACAAAUACG-------AUUCGCUGGCAUCCAG---------CAGUUUGUUUAUGCUUUUGCCCACAGCGGACACAAUUUGUUUUAAUUGCGAGCAAAACA ...((((((((((((((...-------.((((((((((...((---------((.........))))..))))...))))))....)))))).......)))))))).... ( -28.41, z-score = -2.89, R) >droSec1.super_1 388914 95 - 14215200 UCAUUGUUCGCACAAAUACG-------AUUCGCUGGCAUCCAG---------CAGUUUGUUUAUGCUUUCGCCCACAGCGGGCACAAUUUGUUUUAAUUGCGAGCAAAACA ...(((((((((.....(((-------(...(((((...))))---------).................((((.....)))).....))))......))))))))).... ( -28.90, z-score = -2.64, R) >droSim1.chr2R_random 581412 95 - 2996586 UCAUUGUUCGCACAAAUACG-------AUUCGCUGGCAUCCAG---------CAGUUUGUUUAUGCUUUCGCCCACAGCGGGCACAAUUUGUUUUAAUUGCGAGCAAAACA ...(((((((((.....(((-------(...(((((...))))---------).................((((.....)))).....))))......))))))))).... ( -28.90, z-score = -2.64, R) >consensus UCAUUGUUCGCACAAAUACG_______AUUCGCUGGCAUCCAG_________CAGUUUGUUUAUGCUUUCGCCCACAGCGGGCACAAUUUGUUUUAAUUGCGAGCAAAACA ...(((((((((....................(((.....)))...........(((((((...((....))....)))))))...............))))))))).... (-14.59 = -14.30 + -0.29)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:06 2011