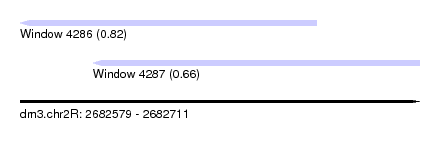
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,682,579 – 2,682,711 |
| Length | 132 |
| Max. P | 0.817076 |

| Location | 2,682,579 – 2,682,677 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 68.05 |
| Shannon entropy | 0.60442 |
| G+C content | 0.52899 |
| Mean single sequence MFE | -35.86 |
| Consensus MFE | -14.53 |
| Energy contribution | -14.25 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.817076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
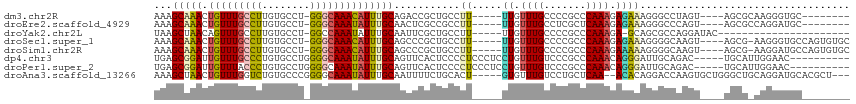
>dm3.chr2R 2682579 98 - 21146708 AAAGCAAACUGUUUGCCUUGUGCCU-GGGCAAACAUUUGCAGACCGCUGCCUU-----UUGUUUGCCCCGCCCAAAGAGAAAGGGCCUAGU----AGCGCAAGGGUGC-------- .((((.....)))).((((((((..-(((((((((...((((....))))...-----.))))))))).((((.........)))).....----.))))))))....-------- ( -40.10, z-score = -2.24, R) >droEre2.scaffold_4929 6776881 98 + 26641161 AAAGCAAACUGUUUGCCUUGUGCCU-GGGCAAAUAUUUGCAACUCGCCGCCUU-----UUGUUUGCCUCGCUCAAAGAGAAAGGGCCCAGU----AGCGCCAGGAUGC-------- .((((.....)))).(((.((((((-(((((((((...((........))...-----.))))).(((..(((...)))..))))))))).----.)))).)))....-------- ( -28.80, z-score = 0.38, R) >droYak2.chr2L 15417564 87 - 22324452 UAAGCUAACAGUUUGCCUUGUGCCU-GGCCAAAUAUUUGCAAUUCGCUGCCUU-----UUGUUUGCCCCGCCCAAAGA-GCAGCGCCAGGAUAC---------------------- (((((.....)))))....((((((-((((((....)))......(((((.((-----(((..((...))..))))).-))))))))))).)))---------------------- ( -23.60, z-score = -0.88, R) >droSec1.super_1 359886 105 - 14215200 AAAGCAAACUGUUUGCCUUGUGCCU-GGGCAAACAUUUGCAGCCCGCUGCCUU-----UUGUUUGCCCCGCCCAAAGAGAAAGGGGCAAGU----AGCG-AAGGGUGCCAGUGUGC ...(((((.(((((((((.......-)))))))))))))).((.(((((((((-----(((((((((((.............)))))))..----.)))-)))))...))))).)) ( -46.02, z-score = -2.71, R) >droSim1.chr2R 1497567 105 - 19596830 AAAGCAAACUGUUUGCCUUGUGCCU-GGGCAAACAUUUGCAGCCCGCUGCCUU-----UUGUUUGCCCCGCCCAAAGAAAAAGGGGCAAGU----AGCG-AAGGAUGCCAGUGUGC ...(((((.(((((((((.......-)))))))))))))).((.(((((((((-----(((((((((((.............)))))))..----.)))-))))..).))))).)) ( -42.72, z-score = -2.41, R) >dp4.chr3 951046 101 - 19779522 UGAGCGGAUUGUUUGCCCUGUGCCUGGGGCAAAUAUUUGCAGUUCACUCCCCUCCCUCCUGUUUGUCCCGCCCAAACAGGGAUUGCAGAC-----UGCAUUGGAAC---------- ...((((..((((((((((......))))))))))(((((((((.............((((((((.......))))))))))))))))))-----)))........---------- ( -35.91, z-score = -2.06, R) >droPer1.super_2 1128799 101 - 9036312 UGAGCGGAUUGUUUACCCUGUGCCUGGGGCAAAUAUUUGCAGUUCACUCCCCUCCCUCCUGUUUGUCCCGCCCAAACAGGGAUUGCAGAC-----UGCAUUGGAAC---------- ...((((.((((...((((((...(((((((((((........................))))))))))).....))))))...)))).)-----)))........---------- ( -31.46, z-score = -1.07, R) >droAna3.scaffold_13266 1578192 106 + 19884421 AAAGCUAACUGUUUGGUCUGUGCCCGGGGCAAAUAUUUGCAAUUUUCUGCACU-----GUGUUUGUCCUGCUCAA--ACACAGGACCAAGUGCUGGGCUGCAGGAUGCACGCU--- ..(((....(((....(((((((((((.((.......((((......))))((-----(((((((.......)))--))))))......)).)))))).)))))..))).)))--- ( -38.30, z-score = -1.63, R) >consensus AAAGCAAACUGUUUGCCUUGUGCCU_GGGCAAAUAUUUGCAGUUCGCUGCCUU_____UUGUUUGCCCCGCCCAAAGAGAAAGGGCCAAGU____AGCG_AAGGAUGC________ ...(((((.(((((((((........))))))))))))))..................((.((((.......)))).))..................................... (-14.53 = -14.25 + -0.28)

| Location | 2,682,603 – 2,682,711 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.43 |
| Shannon entropy | 0.43003 |
| G+C content | 0.50320 |
| Mean single sequence MFE | -40.17 |
| Consensus MFE | -22.50 |
| Energy contribution | -22.24 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.663098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2682603 108 - 21146708 -----GCGGG-AAAAAAGAAAGGUCAAGUGGCAAGGGCUUAAAGCAAACUGUUUGCCUUGUGCCU-GGGCAAACAUUUGCAGACCG-----CUGCCUUUUGUUUGCCCCGCCCAAAGAGA -----(((((-..(((.(((((((..((((((....)).....(((((.(((((((((.......-))))))))))))))....))-----))))))))).)))..)))))......... ( -41.50, z-score = -1.90, R) >droEre2.scaffold_4929 6776905 108 + 26641161 -----GCGGC-AAAAAAGAAAGGUCAGGCGGCAAGGGCUCAAAGCAAACUGUUUGCCUUGUGCCU-GGGCAAAUAUUUGCAACUCG-----CCGCCUUUUGUUUGCCUCGCUCAAAGAGA -----(((((-(((...(((((((..((((((....)).....(((((.(((((((((.......-))))))))))))))....))-----))))))))).))))))..))......... ( -41.00, z-score = -1.76, R) >droYak2.chr2L 15417578 106 - 22324452 -------GCAAAAAAAGAAAAGGUCAACUGGCAAGGGCUUUAAGCUAACAGUUUGCCUUGUGCCU-GGCCAAAUAUUUGCAAUUCG-----CUGCCUUUUGUUUGCCCCGCCCAAAGAG- -------((((((((((....(((((...(((((((((...((((.....))))))))).)))))-))))........(((.....-----.)))))))).))))).............- ( -27.50, z-score = 0.33, R) >droSec1.super_1 359917 109 - 14215200 -----GUGGCAAAAAAAGGAAGGUCAAGUGGCAAGGGCUUAAAGCAAACUGUUUGCCUUGUGCCU-GGGCAAACAUUUGCAGCCCG-----CUGCCUUUUGUUUGCCCCGCCCAAAGAGA -----(.((((((((((((.(((((....)))..(((((....(((((.(((((((((.......-))))))))))))))))))).-----)).)))))).)))))).)........... ( -43.20, z-score = -1.78, R) >droSim1.chr2R 1497598 109 - 19596830 -----GUGGCAAAAAAAGGAAGGUCAAGUGGCAAGGGCUUAAAGCAAACUGUUUGCCUUGUGCCU-GGGCAAACAUUUGCAGCCCG-----CUGCCUUUUGUUUGCCCCGCCCAAAGAAA -----(.((((((((((((.(((((....)))..(((((....(((((.(((((((((.......-))))))))))))))))))).-----)).)))))).)))))).)........... ( -43.20, z-score = -1.94, R) >dp4.chr3 951067 120 - 19779522 GAGCAGGAAAAAAGGGGAGAAGGUCAAGUGGCAAAGGCUUUGAGCGGAUUGUUUGCCCUGUGCCUGGGGCAAAUAUUUGCAGUUCACUCCCCUCCCUCCUGUUUGUCCCGCCCAAACAGG ((((((((....(((((((....((((..(((....)))))))((((((.(((((((((......)))))))))))))))......)))))))...))))))))................ ( -46.80, z-score = -1.69, R) >droPer1.super_2 1128820 118 - 9036312 --AAAAGGAAAAAGGGGAGAAGGUCAAGUGGCAAAGGCUUUGAGCGGAUUGUUUACCCUGUGCCUGGGGCAAAUAUUUGCAGUUCACUCCCCUCCCUCCUGUUUGUCCCGCCCAAACAGG --...(((....(((((((..(.((((..(((....))))))).)(((((((........((((...)))).......))))))).))))))).)))((((((((.......)))))))) ( -40.86, z-score = -0.90, R) >droAna3.scaffold_13266 1578224 109 + 19884421 -----GCGUCGAAAAAGGGAAGGUCAAGUGGCAAAGGCUUAAAGCUAACUGUUUGGUCUGUGCCCGGGGCAAAUAUUUGCAAUUUU-----CUGCACUGUGUUUGUCCUGCUCAAACAC- -----((.........(((.((..((((..(....(((.....)))..)..))))..))...)))(((((((((((.((((.....-----.))))..)))))))))))))........- ( -37.30, z-score = -2.71, R) >consensus _____GCGGCAAAAAAAGGAAGGUCAAGUGGCAAGGGCUUAAAGCAAACUGUUUGCCUUGUGCCU_GGGCAAAUAUUUGCAGUUCG_____CUGCCUUUUGUUUGCCCCGCCCAAAGAGA .....................((.(....((((..(((...((((.....)))))))...))))..(((((((((...(((...........)))....))))))))).).))....... (-22.50 = -22.24 + -0.26)

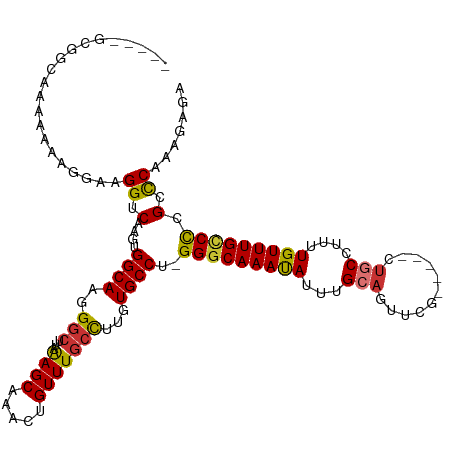
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:06 2011