
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,240,831 – 2,240,921 |
| Length | 90 |
| Max. P | 0.578913 |

| Location | 2,240,831 – 2,240,921 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.92 |
| Shannon entropy | 0.43045 |
| G+C content | 0.43169 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -11.62 |
| Energy contribution | -11.22 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.578913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
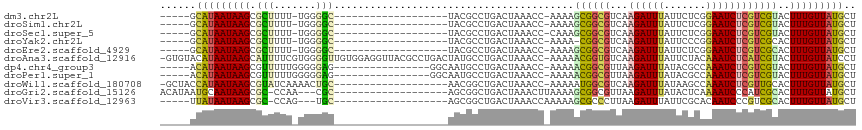
>dm3.chr2L 2240831 90 + 23011544 -----GCAUAAUAAGCGCUUUU-UGGGGC-------------------UACGCCUGACUAAACC-AAAAGCGGCGUCAAGAUUUAUUCUCGGAAUCUCGUCGUACUUUGUUAUGCU -----(((((((((((((((((-(.((((-------------------...)))).).......-)))))))(((.(.((((((.......)))))).).)))..)))))))))). ( -27.01, z-score = -2.48, R) >droSim1.chr2L 2198431 90 + 22036055 -----GCAUAAUAAGCGCUUUU-UGGGGC-------------------UACGCCUGACUAAACC-AAAAGCGGCGUCAAGAUUUAUUCUCGGAAUCUCGUCGUACUUUGUUAUGCU -----(((((((((((((((((-(.((((-------------------...)))).).......-)))))))(((.(.((((((.......)))))).).)))..)))))))))). ( -27.01, z-score = -2.48, R) >droSec1.super_5 415803 90 + 5866729 -----GCAUAAUAAGCGCUUUU-UGGGGC-------------------UACGCCUGACUAAACC-CAAAGCGGCGUCAAGAUUUAUUCUCGGAAUCUCGUCGUACUUUGUUAUGCU -----(((((((((((((((..-((((..-------------------((........))..))-)))))))(((.(.((((((.......)))))).).)))..)))))))))). ( -28.10, z-score = -2.68, R) >droYak2.chr2L 2213958 89 + 22324452 -----GCAUAAUAAGCGCUUUU-UGGGGC-------------------UACGCCUGACUAAACC-AAAA-CGGCGUCAAGAUUUAUUCCCGGAAUCUCGUCGCACUUUGUUAUGCU -----((((((((((.((.(((-((((((-------------------...)))........))-))))-.((((...((((((.......)))))))))))).).))))))))). ( -24.10, z-score = -1.68, R) >droEre2.scaffold_4929 2276211 90 + 26641161 -----GCAUAAUAAGCGCUUUU-UGGGGC-------------------UACGCCUGACUAAACC-AAAAGCGGCGUCAAGAUUUAUUCUCGGAAUCUCGUCGCACUUUGUUAUGCU -----(((((((((((((((((-(.((((-------------------...)))).).......-)))))))(((.(.((((((.......)))))).).)))..)))))))))). ( -29.01, z-score = -2.97, R) >droAna3.scaffold_12916 9952416 114 - 16180835 -GUGUACAUAAUAAGCAUUUUCGUGGGGUUGUGGAGGUUACGCCUGACUAUGCCUGACUAAACC-AAAAACGGUGUCAAGAUUUAUUCUACAAAUCUCAUCGUACUUUGUUAUCCU -............((((....(((((((((((((((((((....))))).....((((...(((-......)))))))........))))).))))))).)).....))))..... ( -22.40, z-score = 0.26, R) >dp4.chr4_group3 7497894 94 - 11692001 -----ACAUAAUAAGCGUUUUUGGGGGAG----------------GGCAAUGCCUGACUAAACC-AAAAACGGCGUUAAGAUUUAUACGCCAAAUCUCGUCGUACUUUGUUAUGCU -----.(((((((((.((((((((((..(----------------(((...))))..))...))-))))))(((((..........)))))..............))))))))).. ( -28.90, z-score = -2.92, R) >droPer1.super_1 4605953 94 - 10282868 -----ACAUAAUAAGCGUUUUUGGGGGAG----------------GGCAAUGCCUGACUAAACC-AAAAACGGCGUUAAGAUUUAUACGCCAAAUCUCGUCGUACUUUGUUAUGCU -----.(((((((((.((((((((((..(----------------(((...))))..))...))-))))))(((((..........)))))..............))))))))).. ( -28.90, z-score = -2.92, R) >droWil1.scaffold_180708 10363978 95 - 12563649 -GCUACCAUAAUAAGCGUAUCAAAACUGC-------------------AACGGCUGACUAAACC-AAAAAUGGCGUCAAGAUUUAUAAGCCAAAUCUCGUUGCACUUUGUUAUGCU -............((((((.((((..(((-------------------((((..((((....((-(....))).))))((((((.......))))))))))))).)))).)))))) ( -24.10, z-score = -3.06, R) >droGri2.scaffold_15126 2205915 93 - 8399593 ACAUAAUGCAAUAAGCGC-CCAA---CGC-------------------AGCGGCUGACUAAACUUAAAAGCGGCGUUAAGAUUUAUACUCAAAAUCCCAUCGCACUUUGUUAUGCU .((((((.......(((.-....---)))-------------------.(((((((.((.........)))))).....(((((.......)))))....))).....)))))).. ( -16.50, z-score = 0.38, R) >droVir3.scaffold_12963 13674850 88 + 20206255 -----UUAUAAUAAGCGC-CCAG---UGC-------------------AGCGGCUGACUAAACCAAAAAGCGCCCUUAAGAUUUAUUCGCACAAUCCCGUCGCACUUUGUUAUGCU -----.((((((((((((-...)---)))-------------------.(((((((.......))....(((...............)))........)))))...)))))))).. ( -15.16, z-score = 0.47, R) >consensus _____GCAUAAUAAGCGCUUUU_UGGGGC___________________UACGCCUGACUAAACC_AAAAGCGGCGUCAAGAUUUAUUCUCGAAAUCUCGUCGUACUUUGUUAUGCU ......(((((((((.(((.......)))........................................((((((...((((((.......))))))))))))..))))))))).. (-11.62 = -11.22 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:34 2011