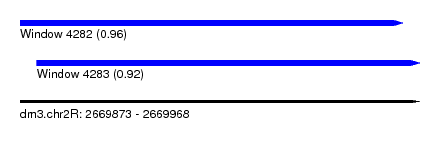
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,669,873 – 2,669,968 |
| Length | 95 |
| Max. P | 0.956509 |

| Location | 2,669,873 – 2,669,964 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 86.93 |
| Shannon entropy | 0.18413 |
| G+C content | 0.38638 |
| Mean single sequence MFE | -24.25 |
| Consensus MFE | -21.94 |
| Energy contribution | -22.27 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
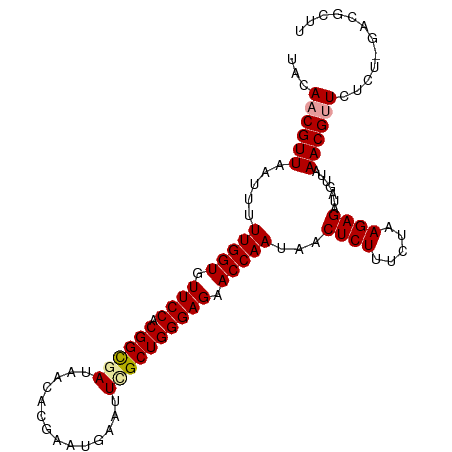
>dm3.chr2R 2669873 91 + 21146708 UACAACGUUAAUGUUUGGUGUUCCACGGCGAUAAAACGAAUGAAUUCGCUGGGAGAACCAAUAACUCUUUCUAAGAGCUAUUUAAACGCUU----------- .....((((.....(((((.((((.((((((..............)))))))))).)))))...((((.....)))).......))))...----------- ( -23.34, z-score = -2.69, R) >droSec1.super_1 341912 102 + 14215200 UACAACGUUAAUUUUUGGUGUUCCACGGCGAUAACACGAAUGAAUUUGCUGGGAGAACCAAUAACUCUUUCUAAGAGAUAGUUAAACGUUCUCUGGACGCUU ................(((((((..((((((...((....))...))))))(((((((...(((((.((((...)))).)))))...)))))))))))))). ( -26.40, z-score = -2.30, R) >droSim1.chr2R 1483931 102 + 19596830 UACAACGUUAAUUUUUGGUGUUCCACGGUGAUAACACGAAUGAAUUGGCUGGGAGAACCAAUAACUCUUUCUAAGAGAUAGUUAAACGUUCUCUUGACGCUU ......((((((((((.(((((..........))))).)).)))))))).((((((((...(((((.((((...)))).)))))...))))))))....... ( -23.00, z-score = -1.10, R) >consensus UACAACGUUAAUUUUUGGUGUUCCACGGCGAUAACACGAAUGAAUUCGCUGGGAGAACCAAUAACUCUUUCUAAGAGAUAGUUAAACGUUCUCU_GACGCUU ...((((((.....(((((.((((.((((((..............)))))))))).)))))...((((.....)))).......))))))............ (-21.94 = -22.27 + 0.34)

| Location | 2,669,877 – 2,669,968 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.64 |
| Shannon entropy | 0.21360 |
| G+C content | 0.39657 |
| Mean single sequence MFE | -24.25 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.34 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.921196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
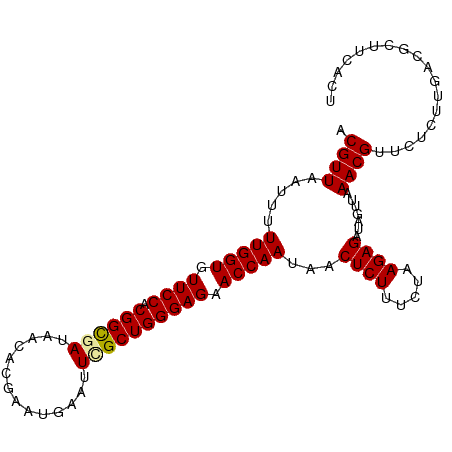
>dm3.chr2R 2669877 91 + 21146708 ACGUUAAUGUUUGGUGUUCCACGGCGAUAAAACGAAUGAAUUCGCUGGGAGAACCAAUAACUCUUUCUAAGAGCUAUUUAAACGCUUCACU----------- .((((.....(((((.((((.((((((..............)))))))))).)))))...((((.....)))).......)))).......----------- ( -23.34, z-score = -2.48, R) >droSec1.super_1 341916 102 + 14215200 ACGUUAAUUUUUGGUGUUCCACGGCGAUAACACGAAUGAAUUUGCUGGGAGAACCAAUAACUCUUUCUAAGAGAUAGUUAAACGUUCUCUGGACGCUUCACU ............(((((((..((((((...((....))...))))))(((((((...(((((.((((...)))).)))))...))))))))))))))..... ( -26.40, z-score = -2.06, R) >droSim1.chr2R 1483935 102 + 19596830 ACGUUAAUUUUUGGUGUUCCACGGUGAUAACACGAAUGAAUUGGCUGGGAGAACCAAUAACUCUUUCUAAGAGAUAGUUAAACGUUCUCUUGACGCUUCACU ..((((((((((.(((((..........))))).)).)))))))).((((((((...(((((.((((...)))).)))))...))))))))........... ( -23.00, z-score = -0.90, R) >consensus ACGUUAAUUUUUGGUGUUCCACGGCGAUAACACGAAUGAAUUCGCUGGGAGAACCAAUAACUCUUUCUAAGAGAUAGUUAAACGUUCUCUUGACGCUUCACU .((((.....(((((.((((.((((((..............)))))))))).)))))...((((.....)))).......)))).................. (-21.34 = -21.34 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:04:02 2011