| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,656,271 – 2,656,380 |
| Length | 109 |
| Max. P | 0.836010 |

| Location | 2,656,271 – 2,656,380 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.00 |
| Shannon entropy | 0.24533 |
| G+C content | 0.36702 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -18.52 |
| Energy contribution | -18.60 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.836010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2656271 109 + 21146708 CUCAUGGAAACCACAUAAAU----AUUUUGUAAU-UCCCCUGACGGCCUCUGGAGCAGCAAAUUAACUCAUUUGUCGUCGUCAAUGUCAGGA----CAUUACAUUUGUGUUAUUUAUG .....(....)..(((((((----(...((((((-...(((((((.....(((.((.((((((......)))))).))..))).))))))).----.)))))).......)))))))) ( -23.20, z-score = -1.12, R) >droSec1.super_1 328404 109 + 14215200 CUCAUGGAAAGCACAUAAAU----AUUUUGUAAU-UCCCCUGACGGCCUCUGGAGCAGCAAAUUAACUCAUUUGUCGUCGUCAAUGUCAGGA----CAUUACAUUUGUGUUAUUUAUG ..((((((.((((((.....----....((((((-...(((((((.....(((.((.((((((......)))))).))..))).))))))).----.))))))..)))))).)))))) ( -26.20, z-score = -1.91, R) >droSim1.chr2R 1470180 109 + 19596830 CUCAUGGAAACCACAUAAAU----AUUUUGUAAU-UCCCCUGACGGCCUCUGGAGCAGCAAAUUAACUCAUUUGUCGUCGUCAAUGUCAGGA----CAUUACAUUUGUGUUAUUUAUG .....(....)..(((((((----(...((((((-...(((((((.....(((.((.((((((......)))))).))..))).))))))).----.)))))).......)))))))) ( -23.20, z-score = -1.12, R) >droYak2.chr2L 15391920 105 + 22324452 ----UGGAAACCACAUAAAU----AUUUUGUAAU-UCCCCUGACGGCCUCUGGAGCAGCAAAUUAACUCAUUUGUCGUCGUCUAUGUCAGGA----CAUUACAUUUUUGUUAUUUAUG ----.(....)..(((((((----(...((((((-...(((((((.....((((((.((((((......)))))).))..))))))))))).----.)))))).......)))))))) ( -23.70, z-score = -1.98, R) >droEre2.scaffold_4929 6751034 105 - 26641161 ----UGAAAGCCACAUAAAU----AUUUUGUAAU-UCCCUUGACGGCCUCUGGAGCAGCAAAUUAACUCAUUUGUCGUCGUCCAUGUCAGGA----CAUUACAUUGUUGUUAUUUAUG ----.........(((((((----(...((((((-......((((((.....(((...........)))....))))))((((......)))----))))))).......)))))))) ( -23.50, z-score = -1.58, R) >droAna3.scaffold_13266 1544630 109 - 19884421 CUCAUGAAUAACACAUAAAU----AUUUUGUAAU-UCCCUUGACGGCUCCUGGAGCAGCAAAUUAACUCAUUUGUCGUCGUUAAUGUCAGGA----CAUAACAUUUAUAAUAUUUAUG .............(((((((----((..(((((.-......((((((((...)))).((((((......))))))))))((((.(((....)----))))))..)))))))))))))) ( -21.90, z-score = -2.03, R) >droWil1.scaffold_180745 2696736 118 - 2843958 CUCAUAAAUAUCUCAUAAAUGCGUAUUAUGUAAGGUCCCUUGACAGUUCUUGAAGCAGCAAAUUAACUCAUUUGUCGUCGUCAAUGUCAGGUAACCUAUUUAAUUUAUGUGAUUUAUG ..(((((((....(((((((((((...)))).((((..(((((((....((((.((.((((((......)))))).))..)))))))))))..)))).....)))))))..))))))) ( -28.90, z-score = -3.88, R) >consensus CUCAUGGAAACCACAUAAAU____AUUUUGUAAU_UCCCCUGACGGCCUCUGGAGCAGCAAAUUAACUCAUUUGUCGUCGUCAAUGUCAGGA____CAUUACAUUUGUGUUAUUUAUG .............(((((((........((((((....(((((((.....(((....((((((......))))))..)))....)))))))......))))))........))))))) (-18.52 = -18.60 + 0.08)

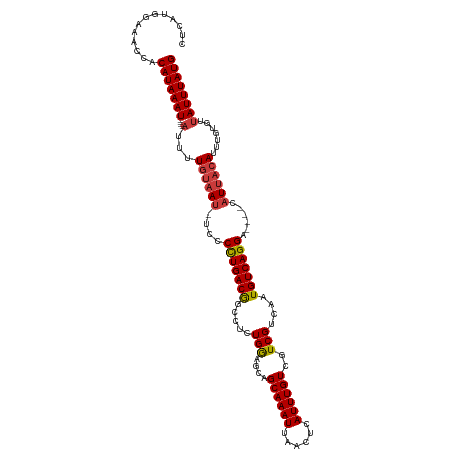

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:59 2011