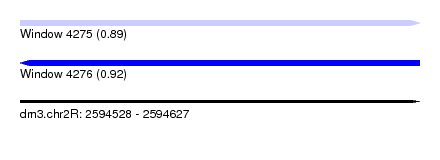
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,594,528 – 2,594,627 |
| Length | 99 |
| Max. P | 0.915746 |

| Location | 2,594,528 – 2,594,627 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 65.54 |
| Shannon entropy | 0.55447 |
| G+C content | 0.33873 |
| Mean single sequence MFE | -19.15 |
| Consensus MFE | -9.20 |
| Energy contribution | -8.63 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.890856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2594528 99 + 21146708 CAGUUAUUAG--AAC------UCAACUGAGAUGAAAGACCAAUAUCGAAAGCCAAAAUUUCCAUGAAAGUGUAGGAUGUUGAUAAGAUUUAAGCUGCACUCCCAUAA (((((.....--..(------((....))).........(((((((((((.......))))(((....)))...)))))))..........)))))........... ( -15.40, z-score = -0.55, R) >droEre2.scaffold_4929 6690194 106 - 26641161 CAGUAAUGAAAAUACCGACUAUUAAAUUUAUUGAAUUAUUAGAGCUCAAAGACAGACAUUCAAUGAAAGCGUAGGAUGCUGAUAAGAUUGAAAUUGCGCUCCCAUA- ((((((....((((.....))))....))))))........((((.(((...(((.(((((.(((....))).))))))))(......)....))).)))).....- ( -15.30, z-score = -0.03, R) >droSec1.super_1 258935 90 + 14215200 CAGUUAUUAGUUAGU------UUAAUAACUAAGAUGAAUGCGAGCUAAUGAGCAAACAUUCCAUGAAAGUGUAGGAUUUU-----------AGCUGCACUCCCAUAU .(((((((((.....------))))))))).....(((((...(((....)))...))))).(((..(((((((......-----------..)))))))..))).. ( -22.30, z-score = -2.85, R) >droSim1.chr2R 1414600 101 + 19596830 CAGUUAUUAGUUAGU------UUAAUAACUAAGAUGAAUGCGAGCUAAAGAGCAAACAUUCCAUGAAAGUGUAGGAUGUUGAUAAGAUUUAAACUGCACUCCCAUAU .(((((((((.....------))))))))).....(((((...(((....)))...))))).(((..(((((((....((((......)))).)))))))..))).. ( -23.60, z-score = -3.04, R) >consensus CAGUUAUUAGUUAAC______UUAAUAACUAAGAAGAAUGAGAGCUAAAAAGCAAACAUUCCAUGAAAGUGUAGGAUGUUGAUAAGAUUUAAACUGCACUCCCAUAU ..............................................................(((..(((((((...................)))))))..))).. ( -9.20 = -8.63 + -0.56)

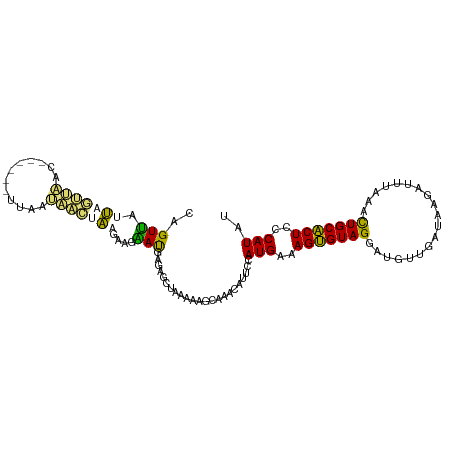
| Location | 2,594,528 – 2,594,627 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 65.54 |
| Shannon entropy | 0.55447 |
| G+C content | 0.33873 |
| Mean single sequence MFE | -19.71 |
| Consensus MFE | -8.30 |
| Energy contribution | -9.73 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.915746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2594528 99 - 21146708 UUAUGGGAGUGCAGCUUAAAUCUUAUCAACAUCCUACACUUUCAUGGAAAUUUUGGCUUUCGAUAUUGGUCUUUCAUCUCAGUUGA------GUU--CUAAUAACUG (((((..((((.((...................)).))))..))))).....((((..((((((...(((.....)))...)))))------)..--))))...... ( -15.41, z-score = 0.30, R) >droEre2.scaffold_4929 6690194 106 + 26641161 -UAUGGGAGCGCAAUUUCAAUCUUAUCAGCAUCCUACGCUUUCAUUGAAUGUCUGUCUUUGAGCUCUAAUAAUUCAAUAAAUUUAAUAGUCGGUAUUUUCAUUACUG -(((.(((((.(((.((((((......(((.......)))...)))))).........))).))))).)))...................(((((.......))))) ( -14.10, z-score = 0.41, R) >droSec1.super_1 258935 90 - 14215200 AUAUGGGAGUGCAGCU-----------AAAAUCCUACACUUUCAUGGAAUGUUUGCUCAUUAGCUCGCAUUCAUCUUAGUUAUUAA------ACUAACUAAUAACUG .((((..((((.((..-----------......)).))))..))))((((((..(((....)))..))))))....(((((((((.------......))))))))) ( -25.30, z-score = -4.61, R) >droSim1.chr2R 1414600 101 - 19596830 AUAUGGGAGUGCAGUUUAAAUCUUAUCAACAUCCUACACUUUCAUGGAAUGUUUGCUCUUUAGCUCGCAUUCAUCUUAGUUAUUAA------ACUAACUAAUAACUG .((((..((((.((...................)).))))..))))((((((..(((....)))..))))))....(((((((((.------......))))))))) ( -24.01, z-score = -4.08, R) >consensus AUAUGGGAGUGCAGCUUAAAUCUUAUCAACAUCCUACACUUUCAUGGAAUGUUUGCCCUUUAGCUCGCAUUCAUCAUAGUAAUUAA______ACUAACUAAUAACUG .((((..((((.((...................)).))))..))))((((((..(((....)))..))))))................................... ( -8.30 = -9.73 + 1.44)

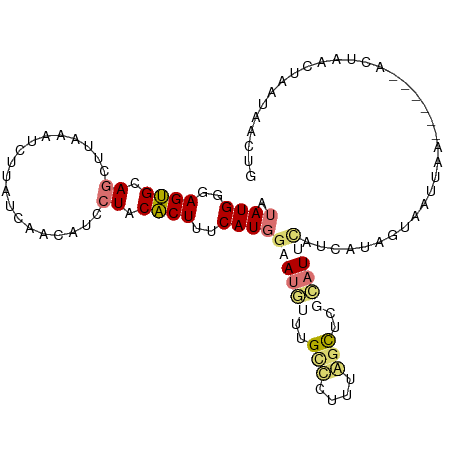
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:56 2011