| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,542,668 – 2,542,733 |
| Length | 65 |
| Max. P | 0.549179 |

| Location | 2,542,668 – 2,542,733 |
|---|---|
| Length | 65 |
| Sequences | 12 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 77.39 |
| Shannon entropy | 0.47340 |
| G+C content | 0.43077 |
| Mean single sequence MFE | -16.71 |
| Consensus MFE | -6.83 |
| Energy contribution | -6.16 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2542668 65 + 21146708 UAUUGCAGUUUGACGUGGAUAACUUCCACAGCGAGGAAGUUAUUACAGAUUGGCUUAAGUAGCAG----- .....(((((((.....((((((((((.......)))))))))).)))))))(((.....)))..----- ( -19.50, z-score = -3.42, R) >droSim1.chr2R 1366848 65 + 19596830 UAUUGCAGUUUGACGUGGAUAACUUCCACAGCGAGGAAGUUAUUACAGAUUGGCUUAAGUAGCAG----- .....(((((((.....((((((((((.......)))))))))).)))))))(((.....)))..----- ( -19.50, z-score = -3.42, R) >droSec1.super_1 209353 65 + 14215200 UAUUGCAGUUUGACGUGGAUAACUUCCACAGCGAGGAAGUUAUUACGGAUUGGCUUAAGUAGCAG----- .....(((((((.....((((((((((.......)))))))))).)))))))(((.....)))..----- ( -18.70, z-score = -2.85, R) >droYak2.chr2L 15287475 65 + 22324452 UAUUCCAGUUUGACGUGGAUAACUUCCACAGCGAGGAAGUUAUUACAGAUUGGCUUAAGUAGCAG----- ((..((((((((.....((((((((((.......)))))))))).))))))))..))........----- ( -19.70, z-score = -3.69, R) >droEre2.scaffold_4929 6638088 65 - 26641161 UAUGGCAGUUUGACGUGGAUAACUUCCACAGCGAGGAAGUUAUUACAGAUUGGCUUAAGUAGCAG----- .....(((((((.....((((((((((.......)))))))))).)))))))(((.....)))..----- ( -19.50, z-score = -3.30, R) >droAna3.scaffold_13266 1396300 64 - 19884421 CUUUUCAGUUUGUUGUGGACAACUUUCACAGCGAGGAAAUCAUAACGGAUUGGCUAAAGUAACA------ ((((((((((((((((((....(((.......)))....))))))))))))))..)))).....------ ( -14.30, z-score = -1.58, R) >dp4.chr3 17284200 63 + 19779522 --UCUCAGUUCGAUGUGGAUAACUUCGACGGCGAGGAAAUCAUCAGUGAUUGGCUGAAAUAGGAG----- --..((((((((.(((.((.....)).))).)))...(((((....))))).)))))........----- ( -13.60, z-score = -0.12, R) >droPer1.super_31 428243 63 - 935084 --UCUCAGUUCGAUGUGGAUAACUUCCACAGCAAGGAAAUCAUCAGUGAUUGGCUGAAAUAGGAG----- --..((((((.(.((((((.....)))))).).....(((((....)))))))))))........----- ( -15.70, z-score = -1.29, R) >droWil1.scaffold_180699 1485061 70 - 2593675 UUUUUCAGUUUGAUGUGGAAAAUCUUCACACCGAAGAGAUCAUCACUGAUUGGCUUACAUGAGAUUGGAA ..(((((((((.((((((....(((((.....)))))(((((....)))))...)))))).))))))))) ( -18.90, z-score = -1.99, R) >droVir3.scaffold_12875 8057691 62 + 20611582 UUUUGCAGUUCGAUGUGGAGACCUUUCACAACGAGGAGAUCAUCACCGACUGGCUAAAGUAA-------- .....(((((((.(((((((...))))))).)))((.(.....).)).))))..........-------- ( -12.40, z-score = -0.00, R) >droMoj3.scaffold_6496 7662179 60 - 26866924 UAUUGCAGUUUGACGUGGAGACAUUUCACAGCGAGGAGAUCAUCACCGACUGGCUAAAGU---------- ....((....(((.(((....))).)))(((((..((.....))..)).)))))......---------- ( -14.90, z-score = -1.39, R) >droGri2.scaffold_15245 1547073 65 + 18325388 UCUUGCAGUUCGAUGUGGAAACCUUUCACAACGAGGAGAUCAUCACCGACUGGCUGAAGUAGUGG----- .(((.(((((((.((((((.....)))))).)))((.(.....).)).....)))))))......----- ( -13.80, z-score = 0.58, R) >consensus UAUUGCAGUUUGACGUGGAUAACUUCCACAGCGAGGAAAUCAUCACAGAUUGGCUUAAGUAGCAG_____ ........((((..(((((.....)))))..))))..((((......))))................... ( -6.83 = -6.16 + -0.67)

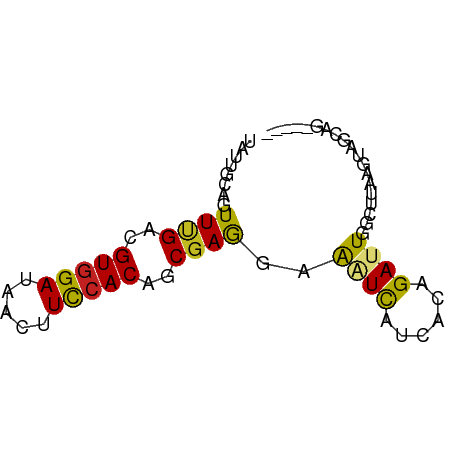
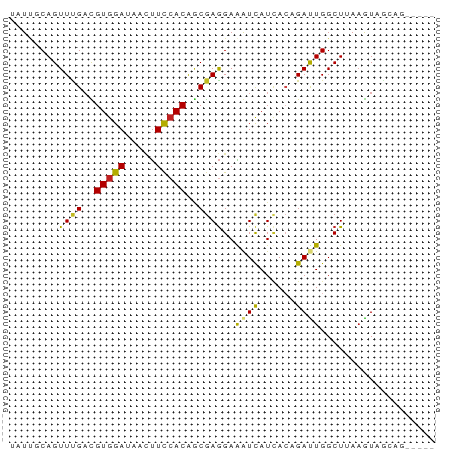
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:51 2011