
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,520,011 – 2,520,146 |
| Length | 135 |
| Max. P | 0.777098 |

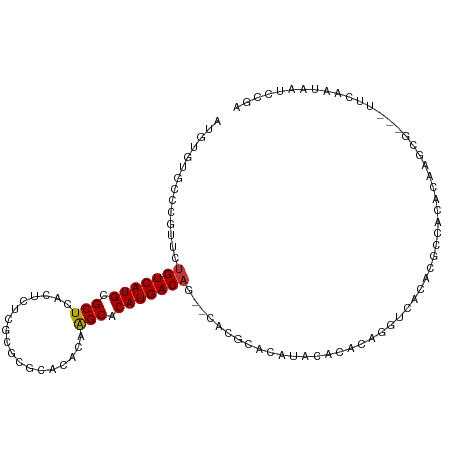
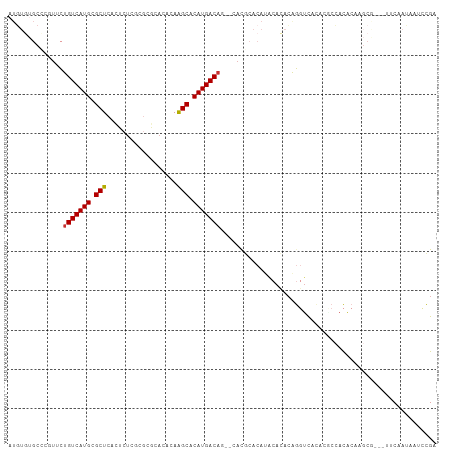
| Location | 2,520,011 – 2,520,108 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 72.24 |
| Shannon entropy | 0.54841 |
| G+C content | 0.50778 |
| Mean single sequence MFE | -22.59 |
| Consensus MFE | -11.04 |
| Energy contribution | -10.93 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2520011 97 - 21146708 AUGUGUGC-----CUGUCAUGCGCUCACUCUCGCGCGCACACAAGCACAUGACAA--CAUGCACAUGCACACAGGUCACACUCCACACAAGCG-----CAAUAAUCCGA ..((((((-----(((((((((((........))))((......))..)))))..--..(((....)))...))).)))))............-----........... ( -26.30, z-score = -0.72, R) >droPer1.super_34 165000 87 - 916997 -----------UCUUGUCAUGCGCUCACGCUCUCACUCACACAGGCACAUGACAU--GACACA-ACGCACUCAACUCUCGCG--AUAAAA------UUCAAUAAUCUGA -----------((.(((((((.(((..................))).))))))).--))....-.(((...........)))--......------............. ( -14.57, z-score = -1.43, R) >dp4.chr3 18221986 87 - 19779522 -----------UCUUGUCAUGCGCUCACGCUCUCACUCACACAGGCACAUGACAU--GACACA-ACGCACUCAACUCUCGCG--AUAAAA------UUCAAUAAUCUGA -----------((.(((((((.(((..................))).))))))).--))....-.(((...........)))--......------............. ( -14.57, z-score = -1.43, R) >droEre2.scaffold_4929 6616016 107 + 26641161 AUGUGUGCCCGUUCUGUCAUGCGCUCACUCUCCCGCGCACACAAGCACAUGACAG--CACGCACAUACACACAGGUCACACACCACACAAGCACAAUCCAAUAAUCCGA .((((((..(((.((((((((.(((..................))).))))))))--.)))..))))))....(((.....)))......................... ( -26.57, z-score = -2.32, R) >droYak2.chr2L 15266193 107 - 22324452 AUGUGUGCCCGUUCUGUCAUGCGCUCACUCUCUCGCGCACACAAGCACAUGACAC--AACGCACAUACACACAGGUCACAUACCACACAAGCGCAAUCCAAUAAUCCGA .((((((..((((.(((((((.(((..................))).))))))).--))))..))))))....(((.....)))......................... ( -24.67, z-score = -1.65, R) >droSec1.super_1 183663 107 - 14215200 AUGUGUGCCCGUUCUGUCAUGCGCUCACUCUCGAGCGCACACAAGCACAUGACAG--CACGCACAUACACACAGGUCACACUCCACACAAGCGCAGUUCAAUAAUCCGA .((((((..(((.((((((((.((((......))))((......)).))))))))--.)))..))))))....((.......))......................... ( -30.30, z-score = -2.00, R) >droSim1.chr2R 1342884 107 - 19596830 AUGUGUGCCCGUUCUGUCAUGCGCUCACUCUCGCGCGCACACAAGCACAUGACAG--CACGCACAUACACACAGGUCACACUCCACACAAGCGCAGUUCAAUAAUCCGA .((((((..(((.(((((((((((........))))((......))..)))))))--.)))..))))))....((.......))......................... ( -28.50, z-score = -1.22, R) >droVir3.scaffold_12875 5134223 101 - 20611582 -UUUAAGCUCAUUCCGUCAUGCGCUCAUACGCACGCACACAUAGGCACAUGACUUCGAACACACAUACAUAAGGGCUCUCAGCGACGCA-------CUCAAAAACUCGA -.....((((.....((((((.((..((............))..)).)))))).....................((.....)))).)).-------............. ( -15.20, z-score = 0.26, R) >consensus AUGUGUGCCCGUUCUGUCAUGCGCUCACUCUCGCGCGCACACAAGCACAUGACAG__CACGCACAUACACACAGGUCACACGCCACACAAGCG___UUCAAUAAUCCGA ..............(((((((.(((..................))).)))))))....................................................... (-11.04 = -10.93 + -0.11)

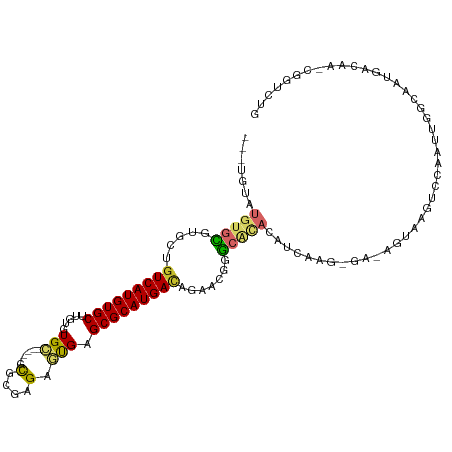
| Location | 2,520,046 – 2,520,146 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 67.81 |
| Shannon entropy | 0.67674 |
| G+C content | 0.49997 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -13.12 |
| Energy contribution | -13.54 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
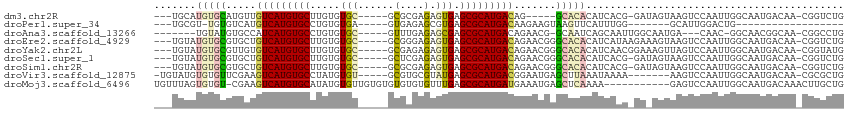
>dm3.chr2R 2520046 100 + 21146708 ---UGCAUGUGCAUGUUGUCAUGUGCUUGUGUGC-----GCGCGAGAGUGAGCGCAUGACAG-----GCACACAUCACG-GAUAGUAAGUCCAAUUGGCAAUGACAA-CGGUCUG ---(((....)))(((((((((((((((((((((-----((((....))..)))))).))))-----)))).......(-(((.....))))........)))))))-))..... ( -40.80, z-score = -1.94, R) >droPer1.super_34 165032 81 + 916997 ---UGCGU-UGUGUCAUGUCAUGUGCCUGUGUGA-----GUGAGAGCGUGAGCGCAUGACAAGAAGUAAGUUCAUUUGG-------GCAUUGGACUG------------------ ---.....-((..((.((((((((((..((((..-----......))))..)))))))))).))..))((((((.....-------....)))))).------------------ ( -23.00, z-score = -0.24, R) >droAna3.scaffold_13266 1369943 97 - 19884421 -------UGUAUGUGCCAUCAUGUGCCUGUGUGC-----GUUUGAGAGCGAGCGCAUGACAGAACG-GCAAUCAGCAAUUGGCAAUGA---CAAC-GGCAACGGCAA-CGGCCUG -------(((...(((((...(((..((((((((-----(((((....))))))))).)))).)))-((.....))...)))))...)---))..-(((..((....-))))).. ( -35.30, z-score = -0.85, R) >droEre2.scaffold_4929 6616056 106 - 26641161 ---UGUAUGUGCGUGCUGUCAUGUGCUUGUGUGC-----GCGGGAGAGUGAGCGCAUGACAGAACGGGCACACAUCAUAAGAAAGUAAGUCCAAUUGGCAAUGACAA-CGGUCUG ---(((.(((.(((.((((((((((((..(...(-----......).)..)))))))))))).))).))).))).............((.((..(((.......)))-.)).)). ( -36.40, z-score = -1.19, R) >droYak2.chr2L 15266233 106 + 22324452 ---UGUAUGUGCGUUGUGUCAUGUGCUUGUGUGC-----GCGAGAGAGUGAGCGCAUGACAGAACGGGCACACAUCAACGGAAAGUUAGUCCAAUUGGCAAUGACAA-CGGUAUG ---(((.(((.((((.(((((((((((..(...(-----....)...)..))))))))))).)))).))).)))....((....((((..((....))...))))..-))..... ( -41.10, z-score = -2.83, R) >droSec1.super_1 183703 105 + 14215200 ---UGUAUGUGCGUGCUGUCAUGUGCUUGUGUGC-----GCUCGAGAGUGAGCGCAUGACAGAACGGGCACACAUCACG-GAUAGUAAGUCCAAUUGGCAAUGACAA-CGGUCUG ---(((.(((.(((.((((((((((((..(...(-----....)...)..)))))))))))).))).))).)))....(-(((.....))))..........(((..-..))).. ( -39.30, z-score = -1.31, R) >droSim1.chr2R 1342924 105 + 19596830 ---UGUAUGUGCGUGCUGUCAUGUGCUUGUGUGC-----GCGCGAGAGUGAGCGCAUGACAGAACGGGCACACAUCACG-GAUAGUAAGUCCAAUUGGCAAUGACAA-CGGUCUG ---(((.(((.(((.((((((((((((..(....-----.(....).)..)))))))))))).))).))).)))....(-(((.....))))..........(((..-..))).. ( -40.00, z-score = -1.20, R) >droVir3.scaffold_12875 5134256 101 + 20611582 -UGUAUGUGUGUUCGAAGUCAUGUGCCUAUGUGU-----GCGUGCGUAUGAGCGCAUGACGGAAUGAGCUUAAAUAAAA-------AAGUCCAAUUGGCAAUGACAA-CGCGCUG -.....(((((((....(((((.((((..(.(((-----.(((((((....)))))))))).).((..(((........-------)))..))...)))))))))))-))))).. ( -32.20, z-score = -1.94, R) >droMoj3.scaffold_6496 1908972 103 - 26866924 UGUUUAGUGUGU-CGAAGUCAUGUGCAUAUGUGUUGUGUGUGUGUGUUUGAGCGCAUGAUGAAAUGAGCUCAAAA-----------GAGUCCAAUUGGCAAUGACAAACUUGCUG .((..(((.(((-((..((((((..(((((.....)))))..))).(((((((.(((......))).))))))).-----------..........)))..))))).))).)).. ( -28.50, z-score = -1.44, R) >consensus ___UGUAUGUGCGUGCUGUCAUGUGCUUGUGUGC_____GCGCGAGAGUGAGCGCAUGACAGAACGGGCACACAUCAAG_GA_AGUAAGUCCAAUUGGCAAUGACAA_CGGUCUG .......(((((.....(((((((((.........................))))))))).......)))))........................................... (-13.12 = -13.54 + 0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:50 2011