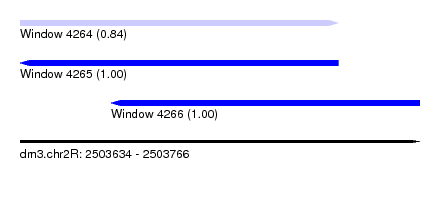
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,503,634 – 2,503,766 |
| Length | 132 |
| Max. P | 0.998946 |

| Location | 2,503,634 – 2,503,739 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Shannon entropy | 0.36479 |
| G+C content | 0.52823 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -15.88 |
| Energy contribution | -16.38 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
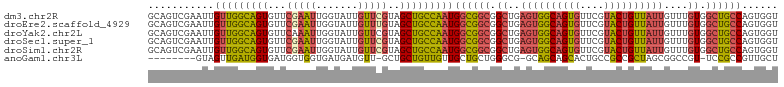
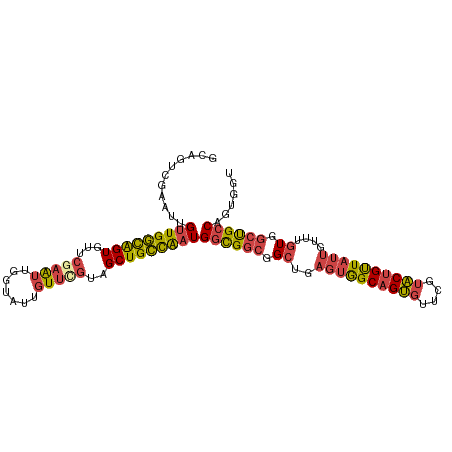
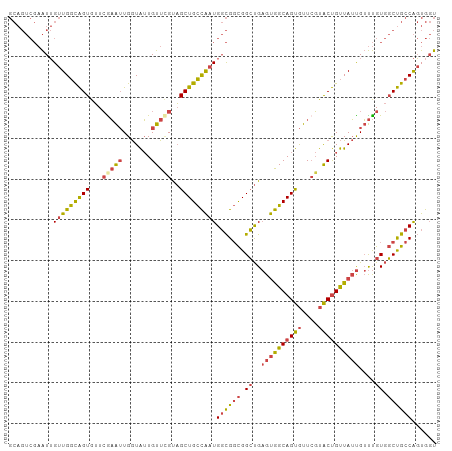
>dm3.chr2R 2503634 105 + 21146708 ACCACUGGCAGCCACAAACAAUAACAGUACGAACACUGCCACUCAGCCGCCGCCAUUGGCAGCUACGAACAAUACCAAUUCGAACACUGCCAACAAUUCGACUGC .....((((((.............((((......))))......(((.((((....)))).))).((((.........))))....))))))............. ( -20.80, z-score = -1.65, R) >droEre2.scaffold_4929 6600119 105 - 26641161 ACCACUGGCAGCCACAAACAAUAACAGUACGAACACUGCCACUCAGCCGCCGCCAUUGGCAGCUACAAACAAUACCAAUUCGAACACUGCCAACAAUUCGACUGC .....((((((.............((((......))))......(((.((((....)))).)))......................))))))............. ( -20.20, z-score = -1.62, R) >droYak2.chr2L 15251329 105 + 22324452 ACCACUGGCAGCCACAAACAAUAACAGUACGAACACUGCCACUCAGCCGCCGCCAUUGGCAGCUACGAACAAUACCAAUUUGAACACUGCCAACAAUUCGACUGC .....((((((...((((......((((......))))......(((.((((....)))).)))..............))))....))))))............. ( -20.80, z-score = -1.50, R) >droSec1.super_1 167364 105 + 14215200 ACCACUGGCAGCCACAAACAAUAACAGUACGAACACUGCCACUCAGCCGCCGCCAUUGGCAGCUACGAACAAUACCAAUUCGAACACUGCCAACAAUUCGACUGC .....((((((.............((((......))))......(((.((((....)))).))).((((.........))))....))))))............. ( -20.80, z-score = -1.65, R) >droSim1.chr2R 1326630 105 + 19596830 ACCACUGGCAGCCACAAACAAUAACAGUACGAACACUGCCACUCAGCCGCCGCCAUUGGCAGCUACGAACAAUACCAAUUCGAACACUGCCAACAAUUCGACUGC .....((((((.............((((......))))......(((.((((....)))).))).((((.........))))....))))))............. ( -20.80, z-score = -1.65, R) >anoGam1.chr3L 7162793 94 + 41284009 AGCAACGGCGGA-ACGGCCGCUAGCGGCGGCAGUGCUGCUGC-CGCCCAGCAGCAACAACAGCAGC-AACAUCAUCACCACCAUCACCAUCAACUAC-------- .((...(((...-...)))(((.((((((((((.....))))-))))..))))).......))...-..............................-------- ( -30.50, z-score = -1.50, R) >consensus ACCACUGGCAGCCACAAACAAUAACAGUACGAACACUGCCACUCAGCCGCCGCCAUUGGCAGCUACGAACAAUACCAAUUCGAACACUGCCAACAAUUCGACUGC .....((((((.............((((......))))......(((.((((....)))).)))......................))))))............. (-15.88 = -16.38 + 0.50)

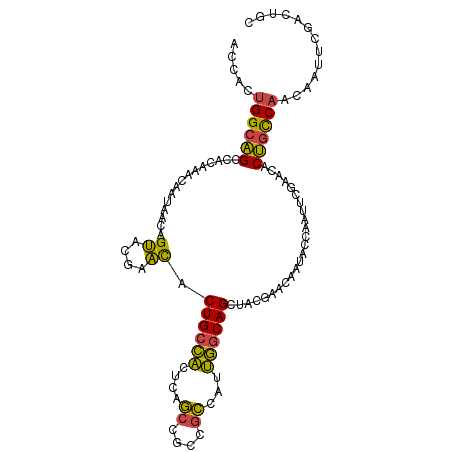
| Location | 2,503,634 – 2,503,739 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.33 |
| Shannon entropy | 0.36479 |
| G+C content | 0.52823 |
| Mean single sequence MFE | -42.78 |
| Consensus MFE | -33.62 |
| Energy contribution | -34.23 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.38 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.998887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2503634 105 - 21146708 GCAGUCGAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUUAUUGUUUGUGGCUGCCAGUGGU ...........(((((((((...(((((.......)))))..)))))))))((((((.((..((((((((((....))))))))))....)).))))))...... ( -43.00, z-score = -3.72, R) >droEre2.scaffold_4929 6600119 105 + 26641161 GCAGUCGAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUUGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUUAUUGUUUGUGGCUGCCAGUGGU ...........(((((((((...(((((.......)))))..)))))))))((((((.((..((((((((((....))))))))))....)).))))))...... ( -40.90, z-score = -3.22, R) >droYak2.chr2L 15251329 105 - 22324452 GCAGUCGAAUUGUUGGCAGUGUUCAAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUUAUUGUUUGUGGCUGCCAGUGGU ((((((((((...((.((((......)))).))..)))))..)))))((.(((((((.((..((((((((((....))))))))))....)).))))))).)).. ( -40.90, z-score = -3.12, R) >droSec1.super_1 167364 105 - 14215200 GCAGUCGAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUUAUUGUUUGUGGCUGCCAGUGGU ...........(((((((((...(((((.......)))))..)))))))))((((((.((..((((((((((....))))))))))....)).))))))...... ( -43.00, z-score = -3.72, R) >droSim1.chr2R 1326630 105 - 19596830 GCAGUCGAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUUAUUGUUUGUGGCUGCCAGUGGU ...........(((((((((...(((((.......)))))..)))))))))((((((.((..((((((((((....))))))))))....)).))))))...... ( -43.00, z-score = -3.72, R) >anoGam1.chr3L 7162793 94 - 41284009 --------GUAGUUGAUGGUGAUGGUGGUGAUGAUGUU-GCUGCUGUUGUUGCUGCUGGGCG-GCAGCAGCACUGCCGCCGCUAGCGGCCGU-UCCGCCGUUGCU --------.........((..(((((((.((((.....-..(((((((((((((....))))-)))))))))..(((((.....))))))))-))))))))..)) ( -45.90, z-score = -2.63, R) >consensus GCAGUCGAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUUAUUGUUUGUGGCUGCCAGUGGU ...........(((((((((...(((((.......)))))..)))))))))((((((.((..((((((((((....))))))))))....)).))))))...... (-33.62 = -34.23 + 0.62)
| Location | 2,503,664 – 2,503,766 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Shannon entropy | 0.36096 |
| G+C content | 0.51161 |
| Mean single sequence MFE | -38.95 |
| Consensus MFE | -30.47 |
| Energy contribution | -32.08 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.998946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
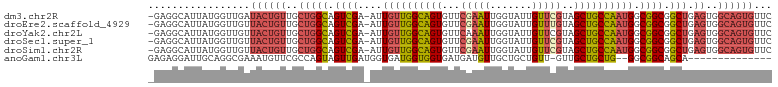
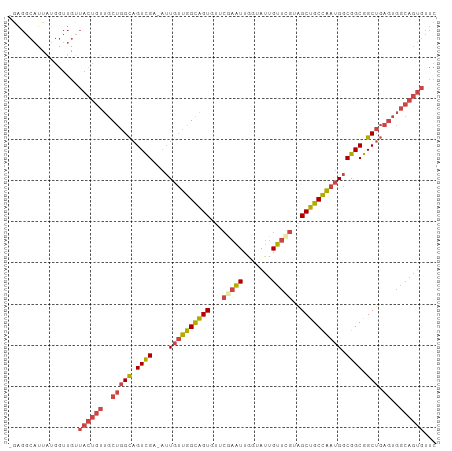
>dm3.chr2R 2503664 102 - 21146708 -GAGGCAUUAUGGUUGAUACUGUUGCUGGCAGUCGA-AUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUC -..............((((((((..(((((.((((.-.(..((((((((...(((((.......)))))..))))))))..))))).))).))..)))))))). ( -43.50, z-score = -4.56, R) >droEre2.scaffold_4929 6600149 102 + 26641161 -GAGGCAUUAUGGUUGUUACUGUUGCUGGCAGUCGA-AUUGUUGGCAGUGUUCGAAUUGGUAUUGUUUGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUC -................((((((..(((((.((((.-.(..((((((((...(((((.......)))))..))))))))..))))).))).))..))))))... ( -39.20, z-score = -3.42, R) >droYak2.chr2L 15251359 102 - 22324452 -GAGGCAUUAUGGUUGUUACUGUUGCUGGCAGUCGA-AUUGUUGGCAGUGUUCAAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUC -................((((((..(((((.((((.-.(..((((((((...(.(((.......))).)..))))))))..))))).))).))..))))))... ( -35.40, z-score = -2.01, R) >droSec1.super_1 167394 102 - 14215200 -GAGGCAUUAUGGUUGUUACUGUUGCUGGCAGUCGA-AUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUC -................((((((..(((((.((((.-.(..((((((((...(((((.......)))))..))))))))..))))).))).))..))))))... ( -41.30, z-score = -3.89, R) >droSim1.chr2R 1326660 102 - 19596830 -GAGGCAUUAUGGUUGUUACUGUUGCUGGCAGUCGA-AUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUC -................((((((..(((((.((((.-.(..((((((((...(((((.......)))))..))))))))..))))).))).))..))))))... ( -41.30, z-score = -3.89, R) >anoGam1.chr3L 7162828 87 - 41284009 GAGAGGAUUGCAGGCGAAAUGUUCGCCAGUAGUUGAUGGUGAUGGUGGUGAUGAUGUUGCUGCUGUU-GUUGCUGCUG--GGCGGCAGCA-------------- .....((((((.(((((.....))))).))))))...((..((((..(..((...))..)..)))).-.))((((((.--...)))))).-------------- ( -33.00, z-score = -2.70, R) >consensus _GAGGCAUUAUGGUUGUUACUGUUGCUGGCAGUCGA_AUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUC .................((((((..((..((((((...(..((((((((...(((((.......)))))..))))))))..)...))))))))..))))))... (-30.47 = -32.08 + 1.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:48 2011