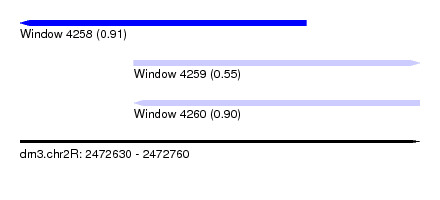
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,472,630 – 2,472,760 |
| Length | 130 |
| Max. P | 0.911220 |

| Location | 2,472,630 – 2,472,723 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 66.92 |
| Shannon entropy | 0.65560 |
| G+C content | 0.61022 |
| Mean single sequence MFE | -22.51 |
| Consensus MFE | -9.43 |
| Energy contribution | -11.03 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
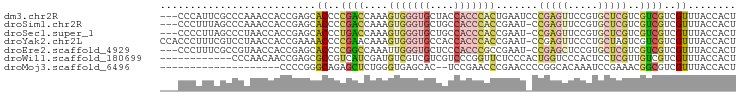
>dm3.chr2R 2472630 93 - 21146708 ---CCCAUUCGCCCAAACCACCGAGCACCCCGACCAAAGUGGGUGCUACCACCCACUGAAUCCCGAGUUCCGUGCUCGUCGUCGUCGUUUACCACU ---...................((((((..((((...((((((((....)))))))).......((((.....))))))))..)).))))...... ( -25.70, z-score = -2.88, R) >droSim1.chr2R 1295079 92 - 19596830 ---CCCUUUAGCCCAAACCACCGAGCACCCCGACCAAAGUGGGUGCUGCCACCCACCGAAU-CCGAGUUCCGUGCUCGUCGUCGUCGUUUACCACU ---...........((((.(((((((((..........(((((((....))))))).((((-....)))).))))))).....)).))))...... ( -24.40, z-score = -2.17, R) >droSec1.super_1 136744 92 - 14215200 ---CCCCUUAGCCCUAACCACCGAGCACCCUGACCAAAGUGGGUGCUGCCACCCACCGAAU-CCGAGUUCCGUGCUCGUCGUCGUCGUUUACCACU ---..................(((((((..........(((((((....))))))).((((-....)))).))))))).................. ( -23.70, z-score = -1.84, R) >droYak2.chr2L 15222025 95 - 22324452 CCACCCUUUCGUCCUAACCACCGAAAACCCCGAACAAAGUGGGUGCCACCACCCACCGAAU-CCGAGUUCCCUGCUAGUCGUCGUCGUUUACCACU ......(((((..........))))).....((((...(((((((....)))))))(((..-...(((.....))).....)))..))))...... ( -18.90, z-score = -1.27, R) >droEre2.scaffold_4929 6568910 92 + 26641161 ---CCCUUUCGCCGUAACCACCGAGCACCCCGGCCAAAUUGGGUGCUCCCACCCGCCGAAU-CCGAGCUCCGUGCUCGUCGUCGUCGUUUACCACU ---..........((((.....((((((((.(......).)))))))).....((.(((..-.(((((.....)))))...))).)).)))).... ( -28.20, z-score = -2.59, R) >droWil1.scaffold_180699 991992 84 + 2593675 ------------CCCAACAACCGAGCGCCGUCAUCGAUGUCGUCGUCGUCCCGGUUCUCCCACUGGUCCCACUCCUCGUUGUCGUCGUUUACCACU ------------....(((((.(((.((((....(((((....)))))...)))).))).....((.......))..))))).............. ( -18.10, z-score = -0.81, R) >droMoj3.scaffold_6496 1845980 74 + 26866924 --------------------CCCCGGGCAGAGCUCUGGGUGAGCAC--UCCGAACCCGAACCCCGGCACAAAUCCGAAACGGCGUCGUUUACCACU --------------------....((((...))))..(((((((((--.(((...(((.....)))..(......)...))).)).)))))))... ( -18.60, z-score = 0.93, R) >consensus ___CCCUUUCGCCCUAACCACCGAGCACCCCGACCAAAGUGGGUGCUGCCACCCACCGAAU_CCGAGUUCCGUGCUCGUCGUCGUCGUUUACCACU ..........................((..((((....(((((((....))))))).......(((((.....)))))..))))..))........ ( -9.43 = -11.03 + 1.60)
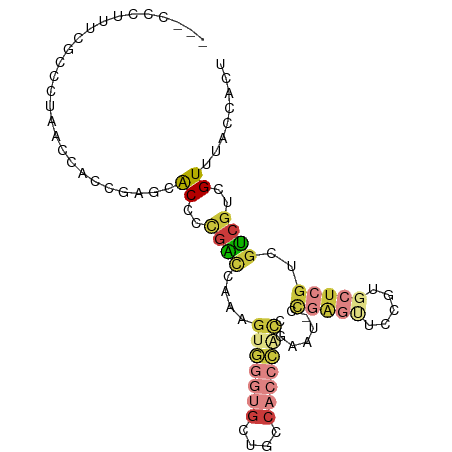
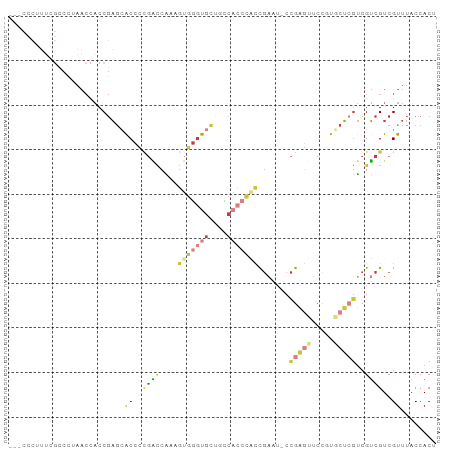
| Location | 2,472,667 – 2,472,760 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 78.55 |
| Shannon entropy | 0.41999 |
| G+C content | 0.60671 |
| Mean single sequence MFE | -38.53 |
| Consensus MFE | -21.72 |
| Energy contribution | -23.42 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.552128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2472667 93 + 21146708 UCAGUGGGUGGUAGCACCCACUUUGGUCGGGGUGCUCGGUGGUUUGGGCGAAUGGGUGGU---GGGAGAUUACCAUCUCCCCUCGGUGUCUGACUG ..((((((((....))))))))..(((((((..((((((....))))))((..(((.(((---((.......))))).))).))....))))))). ( -40.20, z-score = -1.78, R) >droSim1.chr2R 1295115 93 + 19596830 UCGGUGGGUGGCAGCACCCACUUUGGUCGGGGUGCUCGGUGGUUUGGGCUAAAGGGUGGU---GGGAGAUUACCAUCUCCCCUCGGUGUCUGACUG ..((((((((....))))))))..(((((((..((((..((((....))))..))))((.---(((((((....))))))).))....))))))). ( -39.50, z-score = -1.34, R) >droSec1.super_1 136780 93 + 14215200 UCGGUGGGUGGCAGCACCCACUUUGGUCAGGGUGCUCGGUGGUUAGGGCUAAGGGGUGGU---GGGAGAUUACCAUCUCCCCUCGGUGUCUGACUG ..((((((((....))))))))..(((((((..((((........))))..(((((.(((---((.......))))).))))).....))))))). ( -43.50, z-score = -2.28, R) >droYak2.chr2L 15222061 96 + 22324452 UCGGUGGGUGGUGGCACCCACUUUGUUCGGGGUUUUCGGUGGUUAGGACGAAAGGGUGGUGGUGGGAGAUAACCAUCUCCCCUCGGUGUAUGACUG ..((((((((....))))))))....(((((..(((((..........)))))(((.((((((........)))))).)))))))).......... ( -35.00, z-score = -0.70, R) >droEre2.scaffold_4929 6568946 93 - 26641161 UCGGCGGGUGGGAGCACCCAAUUUGGCCGGGGUGCUCGGUGGUUACGGCGAAAGGGUGGU---GGGAGAUUACCAUCUCCCCUCGUUGUAUGAGUG ((((((((((....)))))......)))))...(((((.....((((((((..(((.(((---((.......))))).))).))))))))))))). ( -39.80, z-score = -1.70, R) >droAna3.scaffold_13266 1315429 83 - 19884421 -------------GCGGCGAGUGGGCAGUGGGUGGGUGGUGGCAAGAGCAGUAUGUUGGUGCCGGGAGAUUGCCAUCUCCCCUCGUUGUCUGACUG -------------......(((((((((((((.(((.((((((((...(.((((....))))...)...)))))))).))))))))))))).))). ( -33.20, z-score = -1.08, R) >consensus UCGGUGGGUGGCAGCACCCACUUUGGUCGGGGUGCUCGGUGGUUAGGGCGAAAGGGUGGU___GGGAGAUUACCAUCUCCCCUCGGUGUCUGACUG ..((((((((....))))))))....(((((..((((..(.((....)).)..))))......(((((((....)))))))))))).......... (-21.72 = -23.42 + 1.70)

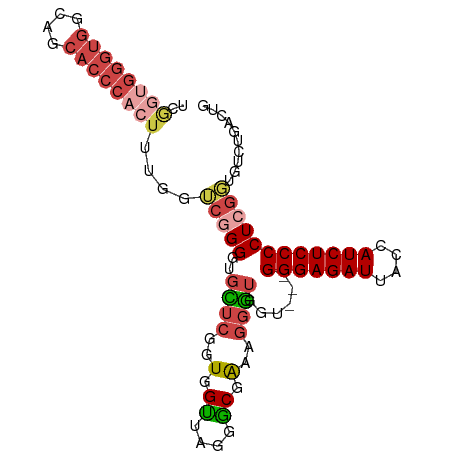
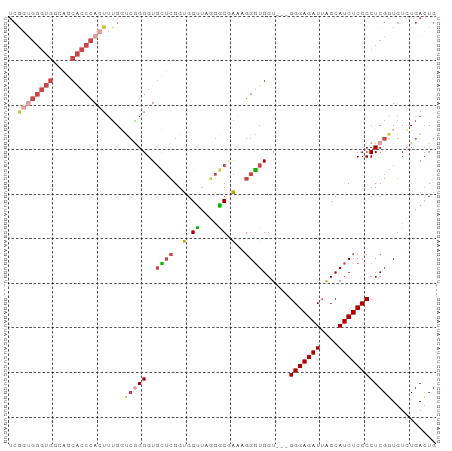
| Location | 2,472,667 – 2,472,760 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.55 |
| Shannon entropy | 0.41999 |
| G+C content | 0.60671 |
| Mean single sequence MFE | -27.81 |
| Consensus MFE | -17.28 |
| Energy contribution | -18.50 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2472667 93 - 21146708 CAGUCAGACACCGAGGGGAGAUGGUAAUCUCCC---ACCACCCAUUCGCCCAAACCACCGAGCACCCCGACCAAAGUGGGUGCUACCACCCACUGA ..(((.......(..(((((((....)))))))---..)........((.(........).)).....)))...((((((((....)))))))).. ( -30.30, z-score = -2.51, R) >droSim1.chr2R 1295115 93 - 19596830 CAGUCAGACACCGAGGGGAGAUGGUAAUCUCCC---ACCACCCUUUAGCCCAAACCACCGAGCACCCCGACCAAAGUGGGUGCUGCCACCCACCGA ..(((.......(..(((((((....)))))))---..)........((.(........).)).....)))....(((((((....)))))))... ( -28.60, z-score = -1.85, R) >droSec1.super_1 136780 93 - 14215200 CAGUCAGACACCGAGGGGAGAUGGUAAUCUCCC---ACCACCCCUUAGCCCUAACCACCGAGCACCCUGACCAAAGUGGGUGCUGCCACCCACCGA ..(((((.....(..(((((((....)))))))---..)........((.(........).))...)))))....(((((((....)))))))... ( -32.10, z-score = -2.65, R) >droYak2.chr2L 15222061 96 - 22324452 CAGUCAUACACCGAGGGGAGAUGGUUAUCUCCCACCACCACCCUUUCGUCCUAACCACCGAAAACCCCGAACAAAGUGGGUGCCACCACCCACCGA ...((.......(..(((((((....)))))))..).......(((((..........))))).....)).....(((((((....)))))))... ( -25.30, z-score = -1.11, R) >droEre2.scaffold_4929 6568946 93 + 26641161 CACUCAUACAACGAGGGGAGAUGGUAAUCUCCC---ACCACCCUUUCGCCGUAACCACCGAGCACCCCGGCCAAAUUGGGUGCUCCCACCCGCCGA ..(((.......)))(((((((....)))))))---.........(((.((........((((((((.(......).)))))))).....)).))) ( -29.42, z-score = -1.80, R) >droAna3.scaffold_13266 1315429 83 + 19884421 CAGUCAGACAACGAGGGGAGAUGGCAAUCUCCCGGCACCAACAUACUGCUCUUGCCACCACCCACCCACUGCCCACUCGCCGC------------- ...........(((((((((((....)))))))((((...............))))...................))))....------------- ( -21.16, z-score = -0.69, R) >consensus CAGUCAGACACCGAGGGGAGAUGGUAAUCUCCC___ACCACCCUUUCGCCCUAACCACCGAGCACCCCGACCAAAGUGGGUGCUACCACCCACCGA ...............(((((((....)))))))..........................................(((((((....)))))))... (-17.28 = -18.50 + 1.22)
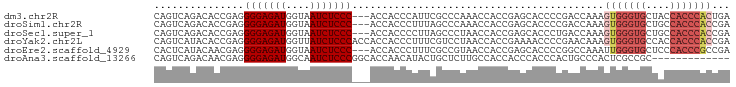
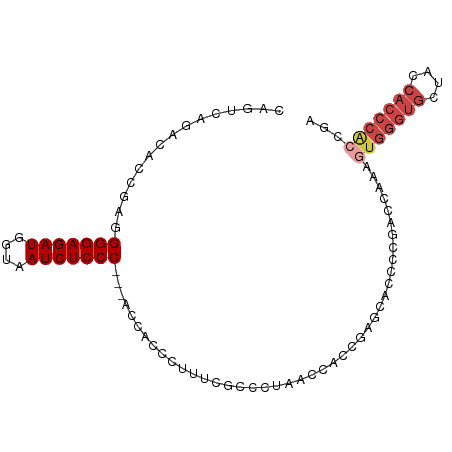
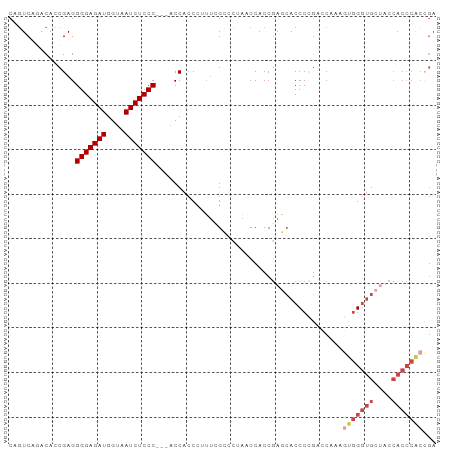
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:43 2011