| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,415,147 – 2,415,268 |
| Length | 121 |
| Max. P | 0.904121 |

| Location | 2,415,147 – 2,415,268 |
|---|---|
| Length | 121 |
| Sequences | 3 |
| Columns | 125 |
| Reading direction | forward |
| Mean pairwise identity | 90.62 |
| Shannon entropy | 0.13022 |
| G+C content | 0.37871 |
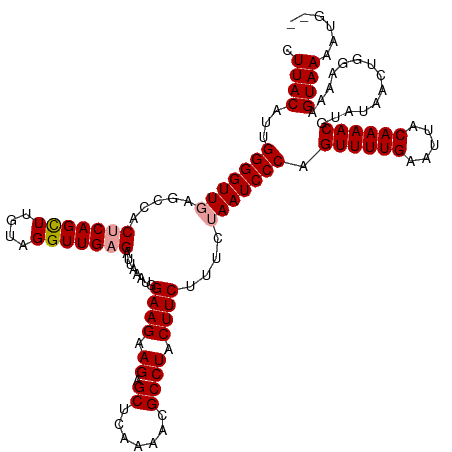
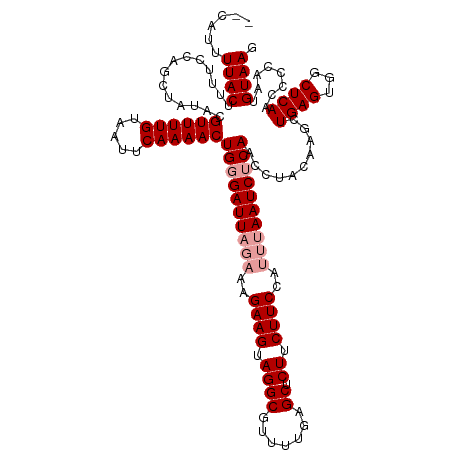
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -25.02 |
| Energy contribution | -25.47 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
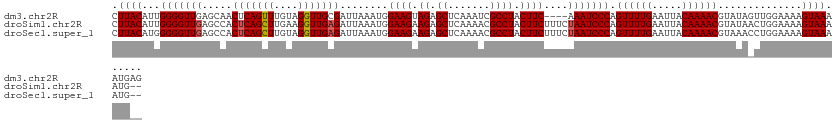
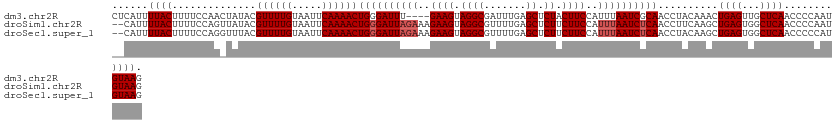
>dm3.chr2R 2415147 121 + 21146708 CUUACAUUGGGGUUGAGCAACUCAGUUUGUAGGUUGCGAUUAAAUGGAAGUAGAGCUCAAAUCGCCUACUUC----AAAUCCCAGUUUUGAAUUACAAAACGUAUAGUUGGAAAAGUAAAAUGAG .((((..(((((((..((((((.........)))))).........(((((((.((.......)))))))))----.)))))))((((((.....))))))..............))))...... ( -32.90, z-score = -2.89, R) >droSim1.chr2R 1241141 123 + 19596830 CUUACAUUGGGGUUGAGCCACUCAGCUUGAAGGUUGAGAUUAAAUGGAAGAAGAGCUCAAAACGCCUACUUCUUUCUAAUCCCAGUUUUGAAUUACAAAACGUAUAACUGGAAAAGUAAAAUG-- .((((..((((((((.....(((((((....))))))).......((((((((.((.......))...))))))))))))))))((((((.....))))))..............))))....-- ( -33.30, z-score = -2.84, R) >droSec1.super_1 85062 123 + 14215200 CUUACAUGGGGGUUGAGCCACUCAGCUUGUAGGUUGAGAUUAAAUGGAAGAAGAGCUCAAAACGCCUACUUCUUUCUAAUCCCAGUUUUGAAUUACAAAACGUAAACCUGGAAAAGUAAAAUG-- .((((...(((((((.....(((((((....))))))).......((((((((.((.......))...))))))))))))))).((((((.....))))))))))..................-- ( -32.50, z-score = -2.09, R) >consensus CUUACAUUGGGGUUGAGCCACUCAGCUUGUAGGUUGAGAUUAAAUGGAAGAAGAGCUCAAAACGCCUACUUCUUUCUAAUCCCAGUUUUGAAUUACAAAACGUAUAACUGGAAAAGUAAAAUG__ .((((...(((((((.....(((((((....)))))))........((((.((.((.......)))).))))....))))))).((((((.....))))))..............))))...... (-25.02 = -25.47 + 0.45)
| Location | 2,415,147 – 2,415,268 |
|---|---|
| Length | 121 |
| Sequences | 3 |
| Columns | 125 |
| Reading direction | reverse |
| Mean pairwise identity | 90.62 |
| Shannon entropy | 0.13022 |
| G+C content | 0.37871 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -20.10 |
| Energy contribution | -21.43 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2415147 121 - 21146708 CUCAUUUUACUUUUCCAACUAUACGUUUUGUAAUUCAAAACUGGGAUUU----GAAGUAGGCGAUUUGAGCUCUACUUCCAUUUAAUCGCAACCUACAAACUGAGUUGCUCAACCCCAAUGUAAG ......((((..............((((((.....))))))((((....----(((((((((.......)).))))))).........((((((........).))))).....))))..)))). ( -28.80, z-score = -3.54, R) >droSim1.chr2R 1241141 123 - 19596830 --CAUUUUACUUUUCCAGUUAUACGUUUUGUAAUUCAAAACUGGGAUUAGAAAGAAGUAGGCGUUUUGAGCUCUUCUUCCAUUUAAUCUCAACCUUCAAGCUGAGUGGCUCAACCCCAAUGUAAG --....((((.....(((((....((((((.....))))))((((((((((..((((.((((.......)).)).))))..)))))))))).......)))))..(((.......)))..)))). ( -28.60, z-score = -2.04, R) >droSec1.super_1 85062 123 - 14215200 --CAUUUUACUUUUCCAGGUUUACGUUUUGUAAUUCAAAACUGGGAUUAGAAAGAAGUAGGCGUUUUGAGCUCUUCUUCCAUUUAAUCUCAACCUACAAGCUGAGUGGCUCAACCCCCAUGUAAG --....((((......(((((...((((((.....)))))).(((((((((..((((.((((.......)).)).))))..))))))))))))))...((((....))))..........)))). ( -29.40, z-score = -1.73, R) >consensus __CAUUUUACUUUUCCAGCUAUACGUUUUGUAAUUCAAAACUGGGAUUAGAAAGAAGUAGGCGUUUUGAGCUCUUCUUCCAUUUAAUCUCAACCUACAAGCUGAGUGGCUCAACCCCAAUGUAAG ......((((..............((((((.....))))))((((((((((..((((.((((.......)).)).))))..))))))))))..........((((...))))........)))). (-20.10 = -21.43 + 1.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:37 2011