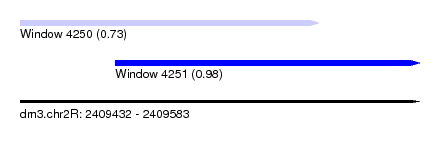
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,409,432 – 2,409,583 |
| Length | 151 |
| Max. P | 0.983897 |

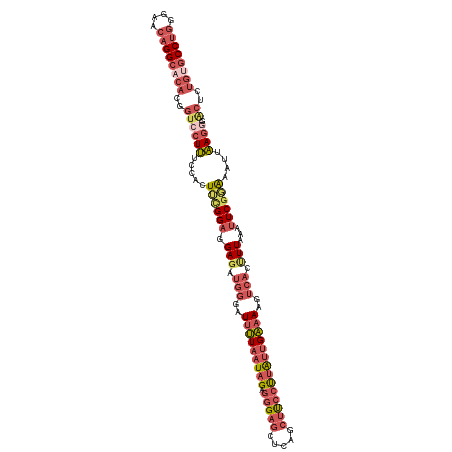
| Location | 2,409,432 – 2,409,545 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.29 |
| Shannon entropy | 0.59410 |
| G+C content | 0.52844 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -17.70 |
| Energy contribution | -19.77 |
| Covariance contribution | 2.07 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 2409432 113 + 21146708 GUGCCUUAAGUGGGUCAAGCUGUGCG--CCAGGGAUGUCCUGGGAACAGGCACACGGUCCUUUUCCCUUUGGAGGAGAUGGGAUUUUAAUAGA-GGGAGCUCAGCUUCCUUAUUGA ((((((..(((.......)))((.(.--(((((.....)))))).))))))))..((((((((((((....).))))).))))))(((((.((-((((((...))))))))))))) ( -39.00, z-score = -0.35, R) >droAna3.scaffold_13266 1256160 97 - 19884421 ---------GUGGCGGAAGUCGCAAAAACUAGGUCAGUCCUGGCGA--GGUAGGGG----UGUUCGCU--GGAGGAGAUGAGUUCCUAAUAGACGGAAACUGCCCGUCCGUGGG-- ---------(((((....)))))...............(((((((.--(((((...----..((((((--(.(((((.....)))))..))).))))..))))))).))).)).-- ( -32.90, z-score = -0.78, R) >droEre2.scaffold_4929 6513412 108 - 26641161 GUGCCUUAAGUGGGUCAAGCUGUGCG--CCAGGAAAACCCUGGAAACAGGCACAGCGUCCUUUCCACUGCGGAGGAUAUGG-AUUUUAAUAG--GGGAGAUAUGCUUCCUUGA--- ...((((.((((((....(((((((.--(((((.....)))))......)))))))......))))))...))))......-.......(((--(((((.....)))))))).--- ( -41.50, z-score = -2.54, R) >droYak2.chr2L 15165643 92 + 22324452 GUGCCUUAAGGAGGUCAAGCUGUGCA--CCAGGAA-AUCCUGGAAACAGGCAAAGUGUCCUUUGCACUGCGGAGGAGAUGGGUUUUCCUUAUU-GA-------------------- ......((((((((.....((((...--(((((..-..)))))..))))((...((.((((((((...)))))))).))..))))))))))..-..-------------------- ( -32.60, z-score = -1.61, R) >droSec1.super_1 79641 113 + 14215200 GUGCCUUAAGUGGGUCAAGCUGUGCG--CCAGGGAUGUCCUGGGAACAGGCACACGGUCCUUUCCACUUCGGAGGAGAUGGGAUUUUAAUAGA-GGGAGCUCAGCUUCCUUAUUGA ...(((((((((((....((((((.(--((.(((....)))(....).))).))))))....)))))))..))))..........(((((.((-((((((...))))))))))))) ( -40.90, z-score = -0.97, R) >droSim1.chr2R 1235197 113 + 19596830 GUGCCUUAAGUGGGUCAAGCUGUGCG--CCAGGGAUGUCCUGGGAACAGGCACACGGUCCUUUCCACUUCGGAGGAGAUGGGAUUUUAAUAGA-GGGAGCUCAGCUUCCUUAUUGA ...(((((((((((....((((((.(--((.(((....)))(....).))).))))))....)))))))..))))..........(((((.((-((((((...))))))))))))) ( -40.90, z-score = -0.97, R) >consensus GUGCCUUAAGUGGGUCAAGCUGUGCG__CCAGGGAUGUCCUGGGAACAGGCACACGGUCCUUUCCACUUCGGAGGAGAUGGGAUUUUAAUAGA_GGGAGCUCAGCUUCCUUAUU__ ...((((.((((((......(((((...(((((.....)))))......)))))........))))))...))))...................(((((.....)))))....... (-17.70 = -19.77 + 2.07)
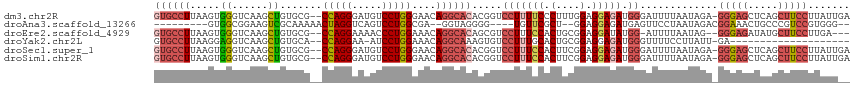
| Location | 2,409,468 – 2,409,583 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.21 |
| Shannon entropy | 0.50742 |
| G+C content | 0.47468 |
| Mean single sequence MFE | -38.16 |
| Consensus MFE | -17.20 |
| Energy contribution | -20.44 |
| Covariance contribution | 3.24 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
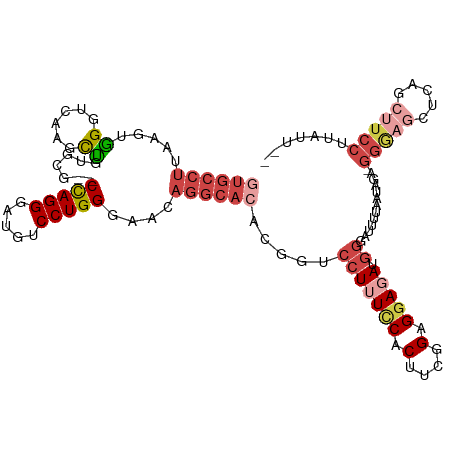
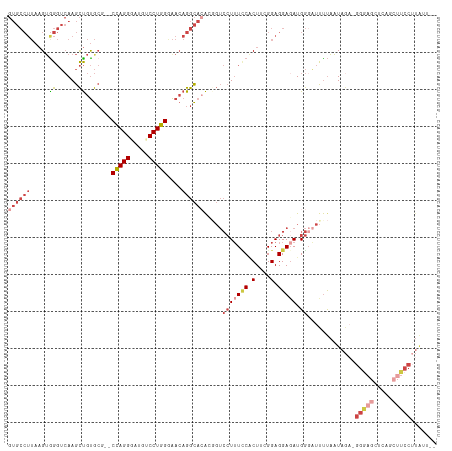
>dm3.chr2R 2409468 115 + 21146708 CCUGGGAACAGGCACACGGUCCUUUUCCCUUUGGAGGAGAUGGGAUUUUAAUAGAGGGAGCUCAGCUUCCUUAUUGAAAAGUCUUUUUAAAUUCGGAAAUUAAGCGAACUCUGUG ((((....))))((((.(((((((((((.....((((((((....(((((((.((((((((...))))))))))))))).))))))))......))))...))).).))).)))) ( -34.80, z-score = -1.30, R) >droAna3.scaffold_13266 1256189 94 - 19884421 CCUGGCGA--GGUAGGGG----UGUUCGCU--GGAGGAGAUGAGUUCCUAAUAGACGGAAA-CUGCCCGUCCGUGGGAACCAGAGCUUGAAGUCCUGAAUUCA------------ .(..((((--(.......----..))))).--.)..(((....(((((((...(((((...-....)))))..)))))))(((..(.....)..)))..))).------------ ( -29.60, z-score = -0.10, R) >droEre2.scaffold_4929 6513448 112 - 26641161 CCUGGAAACAGGCACAGCGUCCUUUCCACUGCGGAGGAUAUGG-AUUUUAAUAG-GGGAGAUAUGCUUCCUUGAUGAAA-GUCAGUUUAACUUCGCAAAUUAAGGGCGCACUGGG ((((....))))....((((((((.....(((((((....((.-.(((((.(((-(((((.....)))))))).)))))-..))......)))))))....))))))))...... ( -45.10, z-score = -5.02, R) >droSec1.super_1 79677 115 + 14215200 CCUGGGAACAGGCACACGGUCCUUUCCACUUCGGAGGAGAUGGGAUUUUAAUAGAGGGAGCUCAGCUUCCUUAUUGAAAAGUCACUUUAAAUUCGGAAAUUAAGGGGACUCUGUG ((((....))))((((.((((((((....((((((.(((.(((..(((((((.((((((((...)))))))))))))))..))).)))...)))))).....)))))))).)))) ( -40.40, z-score = -2.90, R) >droSim1.chr2R 1235233 113 + 19596830 CCUGGGAACAGGCACACGGUCCUUUCCACUUCGGAGGAGAUGGGAUUUUAAUAGAGGGAGCUCAGCUUCCUUAUUGAAAAGUCACUUUAAAUUCGGAAAUUAAGG--ACUCUGUG ((((....))))((((.(((((((.....((((((.(((.(((..(((((((.((((((((...)))))))))))))))..))).)))...))))))....))))--))).)))) ( -40.90, z-score = -3.50, R) >consensus CCUGGGAACAGGCACACGGUCCUUUCCACUUCGGAGGAGAUGGGAUUUUAAUAGAGGGAGCUCAGCUUCCUUAUUGAAAAGUCACUUUAAAUUCGGAAAUUAAGGG_ACUCUGUG ((((....))))((((.(((((((.....((((((.(((.(((..(((((((((.(((((.....))))))))))))))..))).)))...))))))....))))..))).)))) (-17.20 = -20.44 + 3.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:36 2011