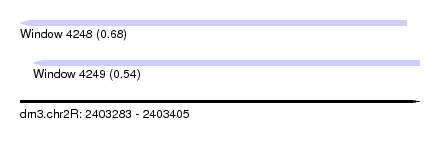
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 2,403,283 – 2,403,405 |
| Length | 122 |
| Max. P | 0.677946 |

| Location | 2,403,283 – 2,403,401 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Shannon entropy | 0.41711 |
| G+C content | 0.54727 |
| Mean single sequence MFE | -38.39 |
| Consensus MFE | -21.62 |
| Energy contribution | -23.38 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.677946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
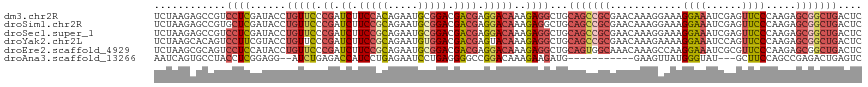
>dm3.chr2R 2403283 118 - 21146708 UCUAAGAGCCGUCCUCGAUACCUGUUCCCGAUCUUCCACAGAAUGCGGACGACGAGGACAAAGAGGCUGCAGCCGCGAACAAAGGAAAGGAAAUCGAGUUCCCAAGAGCGGCUGACUC ......(((((((((((.....(((((.((.(((.....))).)).))))).))))))).....)))).(((((((.......((((..(....)...)))).....))))))).... ( -36.70, z-score = -1.03, R) >droSim1.chr2R 1229172 118 - 19596830 UCUAAGAGCCGUGCUCGAUACCUGUUCCCGAUCUUCCGCAGAAUGCGGACGACGAGGACAAAGAGGCUGCAGCCGCGAACAAAGGAAAGGAAAUCGAGUUCCCAAGAGCGGCUGACUC ......(((((((((((((.(((((((.((.((.(((((.....))))).)))).))))...(.(((....))).)...........)))..)))))))........))))))..... ( -40.60, z-score = -1.65, R) >droSec1.super_1 73962 118 - 14215200 UCUAAGAGCCGUCCUCGAUACCUGUUCCCGAUCUUCCGCAGAAUGCGGACGACGAGGACAAAGAGGCUGCAGCCGCGAACAAAGGAAAGGAAAUCGAGUUCCCAAGAGCGGCUGACUC ......(((((((((((.....((....)).((.(((((.....))))).))))))))).....)))).(((((((.......((((..(....)...)))).....))))))).... ( -40.30, z-score = -1.65, R) >droYak2.chr2L 15159598 118 - 22324452 UCUAAGCACAGUCCUUCGUACCUGUUCCCGAUCUUCCGCAGAAUGUGGACGACGAGUACAAAGAGGCUGCAGCCGCGAACAAAGAAAAGGAAAUCCAGUUCCCAAGAGCGGCUGACUC ........((((((((.((((.(((...((....(((((.....)))))))))).)))).))).)))))(((((((.......(((..((....))..)))......))))))).... ( -37.22, z-score = -2.22, R) >droEre2.scaffold_4929 6507445 118 + 26641161 UCUAAGCGCAGUCCUCCAUACCUGUUCCCGAUCUUCCGCAGAAUGCGGACGACGAGGACAAAGAGGCUGCAGUGGCAAACAAAGCCAAGGAAAUCGCGUUCCCAAGAGCGGCUGACUC ....(((((((.((((......(((((.((.((.(((((.....))))).)))).)))))..))))))))..((((.......)))).........(((((....))))))))..... ( -41.00, z-score = -2.08, R) >droAna3.scaffold_13266 1250150 102 + 19884421 AAUCAGUGCCUACCUCGGAGG--AUCUGAGACCAUCCUGAGAAUCCUGAGGGGCCGGACAAAGAAGAUG-----------GAAGUUAUGGGUAU---GCUUCCAGCCGAGACUGAGUC ..((((((((..((((((.((--.(((.((......)).))).)))))))))))(((..........((-----------(((((.........---))))))).)))..)))))... ( -34.50, z-score = -1.13, R) >consensus UCUAAGAGCCGUCCUCGAUACCUGUUCCCGAUCUUCCGCAGAAUGCGGACGACGAGGACAAAGAGGCUGCAGCCGCGAACAAAGGAAAGGAAAUCGAGUUCCCAAGAGCGGCUGACUC ............((((......(((((.((.((.(((((.....))))).)))).)))))..))))...(((((((............((((......)))).....))))))).... (-21.62 = -23.38 + 1.76)


| Location | 2,403,287 – 2,403,405 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Shannon entropy | 0.42813 |
| G+C content | 0.56239 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -21.05 |
| Energy contribution | -22.33 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
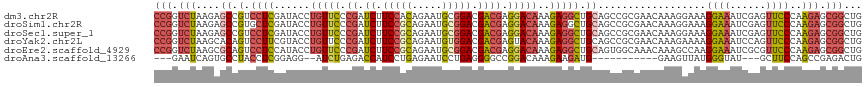
>dm3.chr2R 2403287 118 - 21146708 CCGGUCUAAGAGCCGUCCUCGAUACCUGUUCCCGAUCUUCCACAGAAUGCGGACGACGAGGACAAAGAGGCUGCAGCCGCGAACAAAGGAAAGGAAAUCGAGUUCCCAAGAGCGGCUG .((((((.......(((((((.....(((((.((.(((.....))).)).))))).)))))))....))))))(((((((.......((((..(....)...)))).....))))))) ( -36.80, z-score = -0.72, R) >droSim1.chr2R 1229176 118 - 19596830 CCGGUCUAAGAGCCGUGCUCGAUACCUGUUCCCGAUCUUCCGCAGAAUGCGGACGACGAGGACAAAGAGGCUGCAGCCGCGAACAAAGGAAAGGAAAUCGAGUUCCCAAGAGCGGCUG .((((((..((((...))))......(((((.((.((.(((((.....))))).)))).)))))...))))))(((((((.......((((..(....)...)))).....))))))) ( -42.90, z-score = -1.97, R) >droSec1.super_1 73966 118 - 14215200 CCGGUCUAAGAGCCGUCCUCGAUACCUGUUCCCGAUCUUCCGCAGAAUGCGGACGACGAGGACAAAGAGGCUGCAGCCGCGAACAAAGGAAAGGAAAUCGAGUUCCCAAGAGCGGCUG .((((......)))).((((......(((((.((.((.(((((.....))))).)))).)))))..))))...(((((((.......((((..(....)...)))).....))))))) ( -40.40, z-score = -1.34, R) >droYak2.chr2L 15159602 118 - 22324452 CCGGUCUAAGCACAGUCCUUCGUACCUGUUCCCGAUCUUCCGCAGAAUGUGGACGACGAGUACAAAGAGGCUGCAGCCGCGAACAAAGAAAAGGAAAUCCAGUUCCCAAGAGCGGCUG ............((((((((.((((.(((...((....(((((.....)))))))))).)))).))).)))))(((((((.......(((..((....))..)))......))))))) ( -36.12, z-score = -1.58, R) >droEre2.scaffold_4929 6507449 118 + 26641161 CCGGUCUAAGCGCAGUCCUCCAUACCUGUUCCCGAUCUUCCGCAGAAUGCGGACGACGAGGACAAAGAGGCUGCAGUGGCAAACAAAGCCAAGGAAAUCGCGUUCCCAAGAGCGGCUG .(((((...((((((.((((......(((((.((.((.(((((.....))))).)))).)))))..))))))))..((((.......))))........))((((....))))))))) ( -43.80, z-score = -2.49, R) >droAna3.scaffold_13266 1250154 99 + 19884421 ---GAAUCAGUGCCUACCUCGGAGG--AUCUGAGACCAUCCUGAGAAUCCUGAGGGGCCGGACAAAGAAGAUG-----------GAAGUUAUGGGUAU---GCUUCCAGCCGAGACUG ---....(((((((..((((((.((--.(((.((......)).))).)))))))))))(((..........((-----------(((((.........---))))))).)))..)))) ( -31.60, z-score = -0.75, R) >consensus CCGGUCUAAGAGCCGUCCUCGAUACCUGUUCCCGAUCUUCCGCAGAAUGCGGACGACGAGGACAAAGAGGCUGCAGCCGCGAACAAAGGAAAGGAAAUCGAGUUCCCAAGAGCGGCUG (((.(((....((.(.((((......(((((.((.((.(((((.....))))).)))).)))))..))))).))..................((((......))))..))).)))... (-21.05 = -22.33 + 1.28)
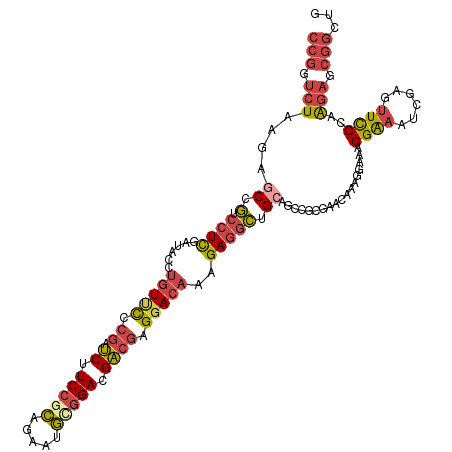
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:03:34 2011